3NX1
 
 | | Crystal structure of Enterobacter sp. Px6-4 Ferulic Acid Decarboxylase | | Descriptor: | Ferulic acid decarboxylase | | Authors: | Gu, W, Yang, J.K, Lou, Z.Y, Meng, Z.H, Zhang, K.-Q. | | Deposit date: | 2010-07-12 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Enzymatic Activity for the Ferulic Acid Decarboxylase (FADase) from Enterobacter sp. Px6-4
Plos One, 6, 2011
|
|
2RJE
 
 | | Crystal structure of L3MBTL1 in complex with H4K20Me2 (residues 17-25), orthorhombic form II | | Descriptor: | CHLORIDE ION, Histone H4, Lethal(3)malignant brain tumor-like protein | | Authors: | Allali-Hassani, A, Liu, Y, Herzanych, N, Ouyang, H, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-14 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | L3MBTL1 recognition of mono- and dimethylated histones.
Nat.Struct.Mol.Biol., 14, 2007
|
|
3NTS
 
 | |
3NV9
 
 | | Crystal structure of Entamoeba histolytica Malic Enzyme | | Descriptor: | ACETATE ION, GLYCEROL, Malic enzyme, ... | | Authors: | Dutta, D, Chakrabarty, A, Ghosh, S.K, Das, A.K. | | Deposit date: | 2010-07-08 | | Release date: | 2011-07-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional characterization of Entamoeba histolytica Malic enzyme
To be Published
|
|
6D54
 
 | |
7AHR
 
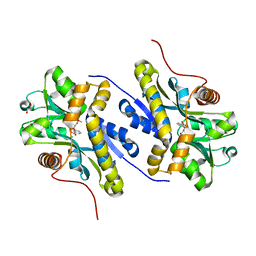 | | Enzyme of biosynthetic pathway | | Descriptor: | 3-[(1-Carboxyvinyl)oxy]benzoic acid, Chorismate dehydratase, GLYCEROL | | Authors: | Archna, A, Breithaupt, C, Stubbs, M.T. | | Deposit date: | 2020-09-25 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Mechanism of chorismate dehydratase MqnA, the first enzyme of the futalosine pathway, proceeds via substrate-assisted catalysis
J.Biol.Chem., 2022
|
|
3O2X
 
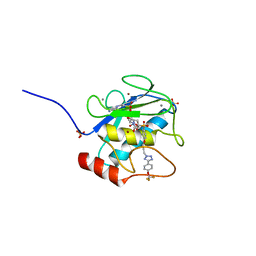 | | MMP-13 in complex with selective tetrazole core inhibitor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, Collagenase 3, ... | | Authors: | Kiefer, J.R, Barta, T.E, Becker, D.P, Mathis, K.J, Williams, J. | | Deposit date: | 2010-07-22 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MMP-13 in complex with selective tetrazole core inhibitor
To be Published
|
|
3NV8
 
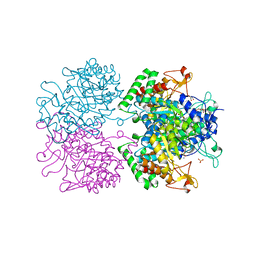 | | The structure of 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase in complex with phosphoenol pyruvate and manganese (thesit-free) | | Descriptor: | CHLORIDE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Parker, E.J, Jameson, G.B, Jiao, W, Hutton, R.H, Webby, C.J, Baker, E.N, Baker, H.M. | | Deposit date: | 2010-07-08 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Synergistic allostery, a sophisticated regulatory network for the control of aromatic amino acid biosynthesis in Mycobacterium tuberculosis
J.Biol.Chem., 285, 2010
|
|
3O4T
 
 | |
3O6I
 
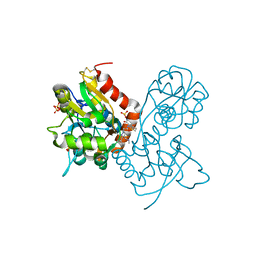 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | 2-[({3-tert-butyl-4-[(methylamino)methyl]-1H-pyrazol-1-yl}acetyl)amino]-4,5,6,7-tetrahydro-1-benzothiophene-3-carboxamide, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Maclean, J.K.F, Campbell, R.A, Cumming, I.A, Gillen, K.J, Gillespie, J, Jamieson, C, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Martin, F, Moir, E.M, Morrow, J.A, Pantling, J, Rankovic, Z, Smith, L. | | Deposit date: | 2010-07-29 | | Release date: | 2010-09-15 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel series of positive modulators of the AMPA receptor: structure-based lead optimization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3NUQ
 
 | | Structure of a putative nucleotide phosphatase from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dong, A, Yang, C, Singer, A.U, Evdokimova, E, Kudritsdka, M, Brown, G, Edwards, A.M, Joachimiak, A, Savchenko, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-07 | | Release date: | 2010-08-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a putative nucleotide phosphatase from Saccharomyces cerevisiae
To be Published
|
|
3NWO
 
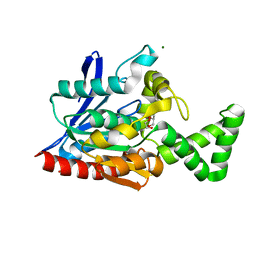 | |
3O03
 
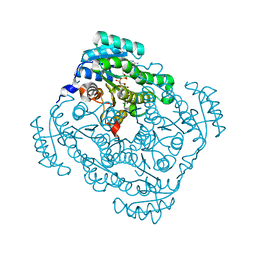 | | Quaternary complex structure of gluconate 5-dehydrogenase from streptococcus suis type 2 | | Descriptor: | CALCIUM ION, D-gluconic acid, Dehydrogenase with different specificities, ... | | Authors: | Peng, H, Gao, F, Zhang, Q, Liu, Y, Gao, G.F. | | Deposit date: | 2010-07-18 | | Release date: | 2010-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insight Into the Catalytic Mechanism of Gluconate 5-Dehydrogenase from Streptococcus Suis: Crystal Structures of the Substrate-Free and Quaternary Complex Enzymes.
Protein Sci., 18, 2009
|
|
3NV5
 
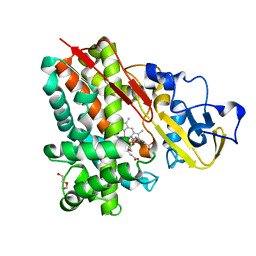 | | Crystal Structure of Cytochrome P450 CYP101D2 | | Descriptor: | Cytochrome P450, DI(HYDROXYETHYL)ETHER, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yang, W, Bell, S.G, Wang, H, Zhou, W.H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-07-08 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The structure of CYP101D2 unveils a potential path for substrate entry into the active site
Biochem.J., 433, 2011
|
|
3OYZ
 
 | | Haloferax volcanii Malate Synthase Pyruvate/Acetyl-CoA Ternary Complex | | Descriptor: | ACETYL COENZYME *A, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Howard, B.R, Bracken, C, Neighbor, A, Thomas, G, Lamlenn, K.K, Schubert, H.L, Whitby, F.G. | | Deposit date: | 2010-09-24 | | Release date: | 2011-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of a halophilic archaeal malate synthase from Haloferax volcanii and comparisons with isoforms A and G.
Bmc Struct.Biol., 11, 2011
|
|
6E21
 
 | | Joint X-ray/neutron structure of PKAc with products Sr2-ADP and phosphorylated peptide SP20 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, STRONTIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A, Gerlits, O.O, Taylor, S. | | Deposit date: | 2018-07-10 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Zooming in on protons: Neutron structure of protein kinase A trapped in a product complex.
Sci Adv, 5, 2019
|
|
3P45
 
 | | Crystal structure of apo-caspase-6 at physiological pH | | Descriptor: | caspase-6 | | Authors: | Mueller, I, Lamers, M.B.A.C, Ritchie, A.J, Dominguez, C, Munoz, I, Maillard, M, Kiselyov, A. | | Deposit date: | 2010-10-06 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal structures of active and inhibitor-bound human Casp6
To be Published
|
|
3OHL
 
 | |
3OHO
 
 | |
6EB3
 
 | | Structural and enzymatic characterization of an esterase from a metagenomic library | | Descriptor: | 1,2-ETHANEDIOL, 1,3-dihydroxypropan-2-yl butanoate, 2-hydroxypropane-1,3-diyl dibutanoate, ... | | Authors: | Guzzo, C.R, Carvalho, C.F, Teixeira, R.D, Farah, C.S. | | Deposit date: | 2018-08-03 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Functional and Structural characterisation of an Esterase from Amazonian Dark Soil
To Be Published
|
|
3P46
 
 | | Integrin binding collagen peptide | | Descriptor: | Synthetic collagen peptide | | Authors: | McEwan, P.A, Emsley, J. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | End-stapled homo and hetero collagen triple helices: a click chemistry approach.
Chem.Commun.(Camb.), 47, 2011
|
|
3PA7
 
 | | Crystal structure of FKBP from plasmodium vivax in complex with tetrapeptide ALPF | | Descriptor: | 4-mer Peptide ALPF, 70 kDa peptidylprolyl isomerase, putative | | Authors: | Balakrishna, A.M, Alag, R, Yoon, H.S. | | Deposit date: | 2010-10-18 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural insights into substrate binding by PvFKBP35, a peptidylprolyl cis-trans isomerase from the human malarial parasite Plasmodium vivax
EUKARYOTIC CELL, 12, 2013
|
|
3M4S
 
 | |
3MC4
 
 | |
3MII
 
 | | Crystal structure of Y0R391Cp/HSP33 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Probable chaperone protein HSP33, SULFATE ION, ... | | Authors: | Guo, P.-C, Zhou, Y.-Y, Zhou, C.-Z, Li, W.-F. | | Deposit date: | 2010-04-10 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Hsp33/YOR391Cp from the yeast Saccharomyces cerevisiae
Acta Crystallogr.,Sect.F, 66, 2010
|
|
