3AAS
 
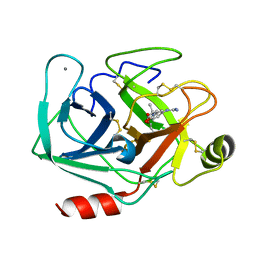 | | Bovine beta-trypsin bound to meta-guanidino schiff base copper (II) chelate | | Descriptor: | (E)-N-[(5-carbamimidamido-2-hydroxyphenyl)methylidene]-L-alanine, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Iyaguchi, D, Kawano, S, Toyota, E. | | Deposit date: | 2009-11-26 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the design of novel Schiff base metal chelate inhibitors of trypsin
Bioorg.Med.Chem., 18, 2010
|
|
3H7D
 
 | | The crystal structure of the cathepsin K Variant M5 in complex with chondroitin-4-sulfate | | Descriptor: | 2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid, CALCIUM ION, Cathepsin K, ... | | Authors: | Cherney, M.M, Kienetz, M, Bromme, D, James, M.N.G. | | Deposit date: | 2009-04-24 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.242 Å) | | Cite: | Structure-activity analysis of cathepsin K/chondroitin 4-sulfate interactions.
J.Biol.Chem., 286, 2011
|
|
3H8C
 
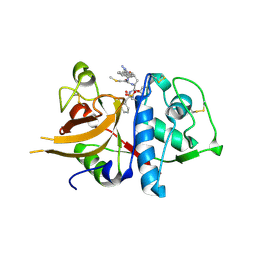 | | A combined crystallographic and molecular dynamics study of cathepsin-L retro-binding inhibitors (compound 14) | | Descriptor: | Cathepsin L1, N-(biphenyl-4-ylacetyl)-S-methyl-L-cysteinyl-D-arginyl-N-(2-phenylethyl)-L-phenylalaninamide | | Authors: | Tulsidas, S.R, Chowdhury, S.F, Kumar, S, Joseph, L, Purisima, E.O, Sivaraman, J. | | Deposit date: | 2009-04-29 | | Release date: | 2009-10-20 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Combined Crystallographic and Molecular Dynamics Study of Cathepsin L Retrobinding Inhibitors
J.Med.Chem., 2009
|
|
2KRG
 
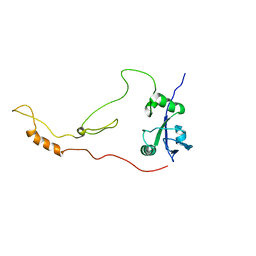 | | Solution Structure of human sodium/ hydrogen exchange regulatory factor 1(150-358) | | Descriptor: | Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Bhattacharya, S, Dai, Z, Li, J, Baxter, S, Callaway, D.J.E, Cowburn, D, Bu, Z. | | Deposit date: | 2009-12-17 | | Release date: | 2009-12-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A conformational switch in the scaffolding protein NHERF1 controls autoinhibition and complex formation.
J.Biol.Chem., 285, 2010
|
|
2K5B
 
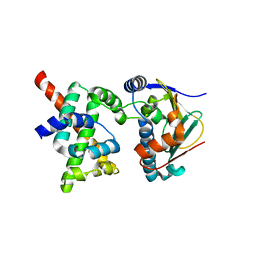 | | Human CDC37-HSP90 docking model based on NMR | | Descriptor: | Heat shock protein HSP 90-alpha, Hsp90 co-chaperone Cdc37 | | Authors: | Sreeramulu, S, Jonker, H.R.A, Lancaster, C.R, Richter, C, Langer, T, Schwalbe, H. | | Deposit date: | 2008-06-26 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The human Cdc37.Hsp90 complex studied by heteronuclear NMR spectroscopy
J.Biol.Chem., 284, 2009
|
|
2K6M
 
 | |
3HB2
 
 | | PrtC methionine mutants: M226I | | Descriptor: | CALCIUM ION, CHLORIDE ION, Secreted protease C, ... | | Authors: | Oberholzer, A.E, Bumann, M, Hege, T, Russo, S, Baumann, U. | | Deposit date: | 2009-05-04 | | Release date: | 2009-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Metzincin's canonical methionine is responsible for the structural integrity of the zinc-binding site
Biol.Chem., 390, 2009
|
|
2KFX
 
 | |
2K7S
 
 | |
2K8F
 
 | | Structural Basis for the Regulation of p53 Function by p300 | | Descriptor: | Cellular tumor antigen p53, Histone acetyltransferase p300 | | Authors: | Bai, Y, Feng, H, Jenkins, L.M, Durell, S.R, Wiodawer, A, Appella, E. | | Deposit date: | 2008-09-08 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for p300 Taz2-p53 TAD1 Binding and Modulation by Phosphorylation.
Structure, 17, 2009
|
|
3EE6
 
 | | Crystal Structure Analysis of Tripeptidyl peptidase -I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Pal, A, Kraetzner, R, Grapp, M, Gruene, T, Schreiber, K, Granborg, M, Urlaub, H, Asif, A.R, Becker, S, Gartner, J, Sheldrick, G.M, Steinfeld, R. | | Deposit date: | 2008-09-04 | | Release date: | 2008-11-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of tripeptidyl-peptidase I provides insight into the molecular basis of late infantile neuronal ceroid lipofuscinosis
J.Biol.Chem., 284, 2009
|
|
2KAY
 
 | | Solution structure and dynamics of S100A5 in the Ca2+ -bound states | | Descriptor: | CALCIUM ION, Protein S100-A5 | | Authors: | Bertini, I, Das Gupta, S, Hu, X, Karavelas, T, Luchinat, C, Parigi, G, Yuan, J, Structural Proteomics in Europe (SPINE), Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2008-11-17 | | Release date: | 2009-06-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of S100A5 in the apo and Ca2+-bound states
J.Biol.Inorg.Chem., 14, 2009
|
|
3ELF
 
 | | Structural Characterization of tetrameric Mycobacterium tuberculosis fructose 1,6-bisphosphate aldolase - substrate binding and catalysis mechanism of a class IIa bacterial aldolase | | Descriptor: | 1,6-FRUCTOSE DIPHOSPHATE (LINEAR FORM), Fructose-bisphosphate aldolase, SODIUM ION, ... | | Authors: | Pegan, S, Rukseree, K, Franzblau, S.G, Mesecar, A.D. | | Deposit date: | 2008-09-22 | | Release date: | 2009-02-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural basis for catalysis of a tetrameric class IIa fructose 1,6-bisphosphate aldolase from Mycobacterium tuberculosis
J.Mol.Biol., 386, 2009
|
|
3COK
 
 | | Crystal structure of PLK4 kinase | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION, Serine/threonine-protein kinase PLK4 | | Authors: | Atwell, S, Burley, S.K, Houle, A, Leon, B, Pelletier, L.A, Sauder, J.M, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-28 | | Release date: | 2008-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of PLK4 kinase.
To be Published
|
|
3EEC
 
 | | X-ray structure of human ubiquitin Cd(II) adduct | | Descriptor: | CADMIUM ION, Ubiquitin | | Authors: | Falini, G, Fermani, S, Tosi, G, Arnesano, F, Natile, G. | | Deposit date: | 2008-09-04 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural probing of Zn(II), Cd(II) and Hg(II) binding to human ubiquitin.
Chem.Commun.(Camb.), 45, 2008
|
|
3CPL
 
 | | Crystal Structure of H-2Db in complex with a variant M6A of the NP366 peptide from influenza A virus | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Kedzierska, K, Guillonneau, C, Hatton, L.A, Stockwell, D, Gras, S, Webby, R, Rossjohn, J, Purcell, A.W, Doherty, P.C, Turner, S.J. | | Deposit date: | 2008-03-31 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Complete modification of TCR specificity and repertoire selection does not perturb a CD8+ T cell immunodominance hierarchy.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3EED
 
 | | Crystal structure of human protein kinase CK2 regulatory subunit (CK2beta; mutant 1-193) | | Descriptor: | Casein kinase II subunit beta, SULFATE ION, ZINC ION | | Authors: | Niefind, K, Raaf, J, Issinger, O.-G. | | Deposit date: | 2008-09-04 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The interaction of CK2{alpha} and CK2{beta}, the subunits of protein kinase CK2, requires CK2{beta} in a preformed conformation and is enthalpically driven
Protein Sci., 17, 2008
|
|
3D06
 
 | | Human p53 core domain with hot spot mutation R249S (I) | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Rozenberg, H, Suad, O, Shimon, L.J.W, Frolow, F, Shakked, Z. | | Deposit date: | 2008-05-01 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis of restoring sequence-specific DNA binding and transactivation to mutant p53 by suppressor mutations
J.Mol.Biol., 385, 2009
|
|
3CS9
 
 | | Human ABL kinase in complex with nilotinib | | Descriptor: | Nilotinib, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Cowan-Jacob, S.W, Fendrich, G, Manley, P, Liebetanz, J, Fabbro, D. | | Deposit date: | 2008-04-09 | | Release date: | 2008-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl
Cancer Cell, 7, 2005
|
|
2KRD
 
 | | Solution Structure of the Regulatory Domain of Human Cardiac Troponin C in Complex with the Switch Region of cardiac Troponin I and W7 | | Descriptor: | CALCIUM ION, N-(6-AMINOHEXYL)-5-CHLORO-1-NAPHTHALENESULFONAMIDE, Troponin C, ... | | Authors: | Oleszczuk, M, Robertson, I.M, Li, M.X, Sykes, B.D. | | Deposit date: | 2009-12-16 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the regulatory domain of human cardiac troponin C in complex with the switch region of cardiac troponin I and W7: the basis of W7 as an inhibitor of cardiac muscle contraction.
J.MOL.CELL.CARDIOL., 48, 2010
|
|
3CTQ
 
 | | Structure of MAP kinase p38 in complex with a 1-o-tolyl-1,2,3-triazole-4-carboxamide | | Descriptor: | Mitogen-activated protein kinase 14, N-benzyl-1-[5-({5-tert-butyl-2-methoxy-3-[(methylsulfonyl)amino]phenyl}carbamoyl)-2-methylphenyl]-1H-1,2,3-triazole-4-carboxamide | | Authors: | Qian, K. | | Deposit date: | 2008-04-14 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based design and subsequent optimization of 2-tolyl-(1,2,3-triazol-1-yl-4-carboxamide) inhibitors of p38 MAP kinase.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3EKL
 
 | | Structural Characterization of tetrameric Mycobacterium tuberculosis fructose 1,6-bisphosphate aldolase - substrate binding and catalysis mechanism of a class IIa bacterial aldolase | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Fructose-bisphosphate aldolase, SODIUM ION, ... | | Authors: | Pegan, S, Rukseree, K, Franzblau, S.G, Mesecar, A.D. | | Deposit date: | 2008-09-19 | | Release date: | 2009-02-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural basis for catalysis of a tetrameric class IIa fructose 1,6-bisphosphate aldolase from Mycobacterium tuberculosis
J.Mol.Biol., 386, 2009
|
|
3D4D
 
 | |
3D4N
 
 | | Crystal Structure of Human 11-beta-Hydroxysteroid Dehydrogenase (HSD1) in Complex with Sulfonamide Inhibitor | | Descriptor: | 1-{[(3R)-3-methyl-4-({4-[(1S)-2,2,2-trifluoro-1-hydroxy-1-methylethyl]phenyl}sulfonyl)piperazin-1-yl]methyl}cyclopropanecarboxamide, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wang, Z, Liu, J, Sudom, A, Walker, N.P. | | Deposit date: | 2008-05-14 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Novel, Potent Benzamide Inhibitors of 11beta-Hydroxysteroid Dehydrogenase Type 1 (11beta-HSD1) Exhibiting Oral Activity in an Enzyme Inhibition ex Vivo Model
J.Med.Chem., 51, 2008
|
|
2KQ6
 
 | | The structure of the EF-hand domain of polycystin-2 suggests a mechanism for Ca2+-dependent regulation of polycystin-2 channel activity | | Descriptor: | Polycystin-2 | | Authors: | Petri, E.T, Celic, A, Kennedy, S.D, Ehrlich, B.E, Boggon, T.J, Hodsdon, M.E. | | Deposit date: | 2009-10-28 | | Release date: | 2010-05-12 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of the EF-hand domain of polycystin-2 suggests a mechanism for Ca2+-dependent regulation of polycystin-2 channel activity.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
