3D12
 
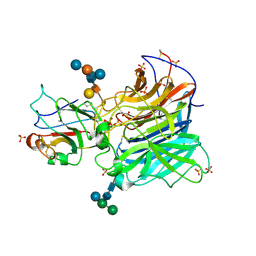 | | Crystal Structures of Nipah Virus G Attachment Glycoprotein in Complex with its Receptor Ephrin-B3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ephrin-B3, ... | | Authors: | Xu, K, Rajashankar, K.R, Chan, Y.P, Himanen, P, Broder, C.C, Nikolov, D.B. | | Deposit date: | 2008-05-02 | | Release date: | 2008-08-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Host cell recognition by the henipaviruses: crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
4P14
 
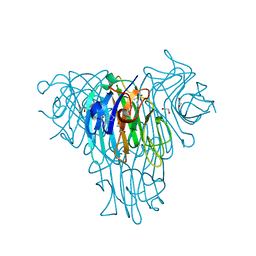 | | Crystal structure of Canavalia brasiliensis (ConBr) complexed with adenine | | Descriptor: | 1,2-ETHANEDIOL, ADENINE, CALCIUM ION, ... | | Authors: | Delatorre, P, Rocha, B.A.M, Silva-Filho, J.C, Teixeira, C.S, Cavada, B.S, Nascimento, K.S, Neto, I.L.B, Nobrega, R.B, Nagano, C.S, Sampaio, A.H, Holanda, E, Leal, R.B. | | Deposit date: | 2014-02-24 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Canavalia brasiliensis (ConBr) complexed with adenine
To Be Published
|
|
6VS6
 
 | |
7AKD
 
 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with the 47D11 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 47D11 neutralizing antibody heavy chain, ... | | Authors: | Fedry, J, Hurdiss, D.L, Wang, C, Li, W, Obal, G, Drulyte, I, Howes, S.C, van Kuppeveld, F.J.M, Foerster, F, Bosch, B.J. | | Deposit date: | 2020-09-30 | | Release date: | 2021-05-19 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the cross-neutralization of SARS-CoV and SARS-CoV-2 by the human monoclonal antibody 47D11.
Sci Adv, 7, 2021
|
|
4P1V
 
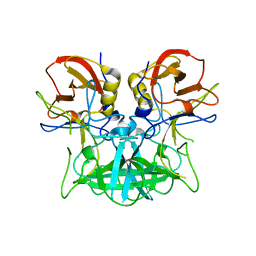 | | Structure of the P domain from a GI.7 Norovirus variant in complex with H-type 2 HBGA | | Descriptor: | P domain of VPI, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-02-27 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5497 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
6P8K
 
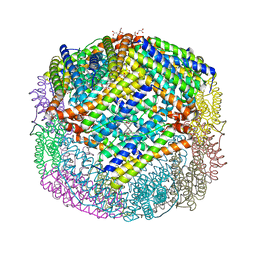 | | Escherichia coli Bacterioferritin Substituted with Zinc Protoporphyrin IX | | Descriptor: | Bacterioferritin, MALONATE ION, PROTOPORPHYRIN IX CONTAINING ZN, ... | | Authors: | Taylor, A.B, Cioloboc, D, Kurtz, D.M. | | Deposit date: | 2019-06-07 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a Zinc Porphyrin-Substituted Bacterioferritin and Photophysical Properties of Iron Reduction.
Biochemistry, 59, 2020
|
|
4PFQ
 
 | | Crystal structure of hypoxanthine phosphoribosyltransferase from Brachybacterium faecium DSM 4810, NYSGRC Target 029763. | | Descriptor: | Hypoxanthine phosphoribosyltransferase, MAGNESIUM ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-04-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of hypoxanthine phosphoribosyltransferase from Brachybacterium faecium DSM 4810, NYSGRC Target 0299763.
to be published
|
|
4PCR
 
 | | Crystal structure of Canavalia brasiliensis seed lectin (ConBr) complexed with Gamma-Aminobutyric Acid (GABA) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Concanavalin-Br, ... | | Authors: | Delatorre, P, Rocha, B.A.M, Silva-Filho, J.C, Teixeira, C.S, Cavada, B.S, Nascimento, K.S, Nbrega, R.B, Nagano, C.S, Sampaio, A.H, Leal, R.B, Neto, I.L.B. | | Deposit date: | 2014-04-16 | | Release date: | 2015-04-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of Canavalia brasiliensis seed lectin (ConBr) complexed with Gamma-Aminobutyric Acid (GABA)
To Be Published
|
|
1EW9
 
 | | ALKALINE PHOSPHATASE (E.C. 3.1.3.1) COMPLEX WITH MERCAPTOMETHYL PHOSPHONATE | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, MERCAPTOMETHYL PHOSPHONATE, ... | | Authors: | Holtz, K.M, Stec, B, Meyers, J.K, Antonelli, S.M, Widlanski, T.S, Kantrowitz, E.R. | | Deposit date: | 2000-04-24 | | Release date: | 2002-05-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Alternate modes of binding in two crystal structures of alkaline phosphatase-inhibitor complexes.
Protein Sci., 9, 2000
|
|
6D0K
 
 | | Crystal structure of a CLC-type fluoride/proton antiporter, E118Q mutant | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, CLC-type fluoride/proton antiporter, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Last, N.B, Stockbridge, R.B, Wilson, A.E, Shane, T, Kolmakova-Partensky, L, Koide, A, Koide, S, Miller, C. | | Deposit date: | 2018-04-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | A CLC-type F-/H+antiporter in ion-swapped conformations.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6CSW
 
 | | Crystal Structure of the Human vaccinia-related kinase bound to a N-methyl-N-propyl-dihydropteridine inhibitor | | Descriptor: | (7R)-5-butyl-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-7,8-dimethyl-7,8-dihydropteridin-6(5H)-one, ACETATE ION, CHLORIDE ION, ... | | Authors: | dos Reis, C.V, de Souza, G.P, Counago, R.M, Chiodi, C.G, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-21 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Human vaccinia-related kinase bound to a N-methyl-N-propyl-dihydropteridine inhibitor
To Be Published
|
|
6CMM
 
 | | Crystal Structure of the Human vaccinia-related kinase bound to a N,N-dipropynyl-dihydropteridine inhibitor | | Descriptor: | (7R)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-7-methyl-5,8-di(prop-2-yn-1-yl)-7,8-dihydropteridin-6(5H)-one, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | dos Reis, C.V, de Souza, G.P, Counago, R.M, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-05 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Human vaccinia-related kinase bound to a N,N-dipropynyl-dihydropteridine inhibitor
To Be Published
|
|
1EHN
 
 | | CRYSTAL STRUCTURE OF CHITINASE A MUTANT E315Q COMPLEXED WITH OCTA-N-ACETYLCHITOOCTAOSE (NAG)8. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | Authors: | Papanikolau, Y, Prag, G, Tavlas, G, Vorgias, C.E, Oppenheim, A.B, Petratos, K. | | Deposit date: | 2000-02-22 | | Release date: | 2001-02-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis.
Biochemistry, 40, 2001
|
|
1O9M
 
 | | The Complex of a novel antibiotic with the Aminoacyl Site of the Bacterial Ribosome Revealed by X-Ray Crystallography. | | Descriptor: | 1-(AMINOETHYL)AMINO-4-AMINOBUTANE, 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranose, 4-AMINO-2-HYDROXYBUTANOIC ACID, ... | | Authors: | Russell, R, Murray, J.B. | | Deposit date: | 2002-12-17 | | Release date: | 2003-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Complex of a Designer Antibiotic with a Model Aminoacyl Site of the 30S Ribosomal Subunit Revealed by X-Ray Crystallography
J.Am.Chem.Soc., 125, 2003
|
|
6P3W
 
 | |
4P1L
 
 | | Crystal structure of a trap periplasmic solute binding protein from chromohalobacter salexigens dsm 3043 (csal_2479), target EFI-510085, with bound d-glucuronate, spg i213 | | Descriptor: | GLYCEROL, SULFATE ION, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-02-26 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
1EVW
 
 | | L116A MUTANT OF THE HOMING ENDONUCLEASE I-PPOI COMPLEXED TO HOMING SITE DNA. | | Descriptor: | DNA (5'-D(*TP*GP*AP*CP*TP*CP*TP*CP*TP*TP*AP*A)-3'), DNA (5'-D(*TP*GP*GP*CP*TP*AP*CP*CP*TP*TP*AP*A)-3'), DNA (5'-D(P*GP*AP*GP*AP*GP*TP*CP*A)-3'), ... | | Authors: | Galburt, E.A, Jurica, M.S, Chevalier, B.S, Erho, D, Stoddard, B.L. | | Deposit date: | 2000-04-20 | | Release date: | 2000-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Conformational changes and cleavage by the homing endonuclease I-PpoI: a critical role for a leucine residue in the active site.
J.Mol.Biol., 300, 2000
|
|
4P3I
 
 | | Structure of the P domain from a GI.7 Norovirus variant in complex with LeA HBGA. | | Descriptor: | P domain of VP1, beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-03-07 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6941 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
5WA9
 
 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant nucleoside D-Ala phosphoramidate substrate complex | | Descriptor: | CHLORIDE ION, Histidine triad nucleotide-binding protein 1, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-~{N}-[(2~{R})-1-methoxy-1-oxidanylidene-propan-2-yl]phosphonamidic acid | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2017-06-26 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | A Crystal Structure Based Guide to the Design of Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) Activated ProTides.
Mol. Pharm., 14, 2017
|
|
1EVK
 
 | | CRYSTAL STRUCTURE OF A TRUNCATED FORM OF THREONYL-TRNA SYNTHETASE WITH THE LIGAND THREONINE | | Descriptor: | THREONINE, THREONYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Sankaranarayanan, R, Dock-Bregeon, A.C, Rees, B, Moras, D. | | Deposit date: | 2000-04-20 | | Release date: | 2000-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Zinc ion mediated amino acid discrimination by threonyl-tRNA synthetase.
Nat.Struct.Biol., 7, 2000
|
|
2RTL
 
 | |
6PEH
 
 | | Crystal structure of rabbit monoclonal anti-HIV antibody 1C2 | | Descriptor: | 1C2 Fab Heavy Chain, 1C2 Fab Light Chain, SULFATE ION | | Authors: | Liban, T, Pancera, M. | | Deposit date: | 2019-06-20 | | Release date: | 2019-11-20 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.296 Å) | | Cite: | Vaccination with Glycan-Modified HIV NFL Envelope Trimer-Liposomes Elicits Broadly Neutralizing Antibodies to Multiple Sites of Vulnerability.
Immunity, 51, 2019
|
|
2C0A
 
 | | Mechanism of the Class I KDPG aldolase | | Descriptor: | KHG/KDPG ALDOLASE, PHOSPHATE ION | | Authors: | Hutchins, M.J, Fullerton, S.W.B, Toone, E.J, Naismith, J.H. | | Deposit date: | 2005-08-29 | | Release date: | 2005-09-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanism of the Class I Kdpg Aldolase.
Bioorg.Med.Chem., 14, 2006
|
|
6CT7
 
 | |
6CDQ
 
 | |
