2HSG
 
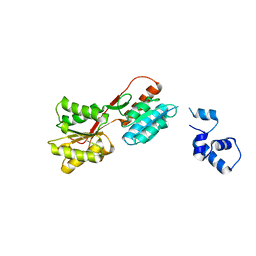 | |
1QXF
 
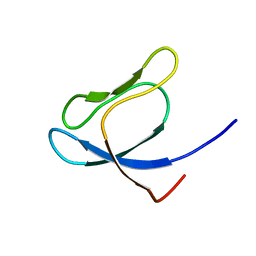 | | SOLUTION STRUCTURE OF 30S RIBOSOMAL PROTEIN S27E FROM ARCHAEOGLOBUS FULGIDUS: GR2, A NESG TARGET PROTEIN | | Descriptor: | 30S RIBOSOMAL PROTEIN S27E | | Authors: | Herve Du Penhoat, C, Atreya, H.S, Shen, Y, Liu, G, Acton, T.B, Xiao, R, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-09-05 | | Release date: | 2003-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of the 30S ribosomal protein S27e encoded in gene RS27_ARCFU of Archaeoglobus fulgidis reveals a novel protein fold
Protein Sci., 13, 2004
|
|
2D46
 
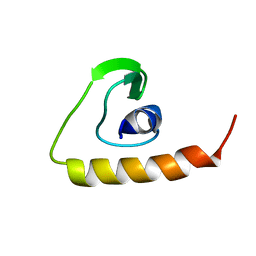 | | Solution Structure of the Human Beta4a-A Domain | | Descriptor: | calcium channel, voltage-dependent, beta 4 subunit isoform a | | Authors: | Vendel, A.C, Rithner, C.D, Lyons, B.A, Horne, W.A. | | Deposit date: | 2005-10-10 | | Release date: | 2005-10-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal A domain of the human voltage-gated Ca2+channel beta4a subunit
Protein Sci., 15, 2006
|
|
1U97
 
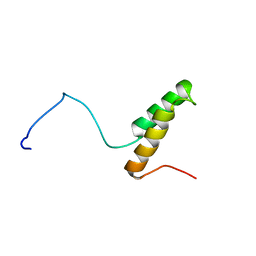 | |
1BY8
 
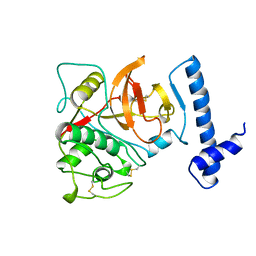 | | THE CRYSTAL STRUCTURE OF HUMAN PROCATHEPSIN K | | Descriptor: | PROTEIN (PROCATHEPSIN K) | | Authors: | Lalonde, J.M, Zhao, B, Smith, W.W, Janson, C.A, Desjarlais, R.L, Tomaszek, T.A, Carr, T.J, Thompson, S.K, Yamashita, D.S, Veber, D.F, Abdel-Mequid, S.S. | | Deposit date: | 1998-10-27 | | Release date: | 1999-10-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of human procathepsin K.
Biochemistry, 38, 1999
|
|
1TRF
 
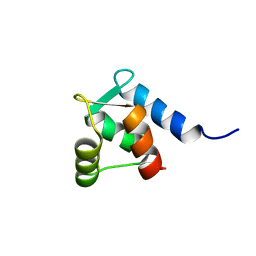 | |
2ND4
 
 | |
3MRA
 
 | | M3 TRANSMEMBRANE SEGMENT OF ALPHA-SUBUNIT OF NICOTINIC ACETYLCHOLINE RECEPTOR FROM TORPEDO CALIFORNICA, NMR, 15 STRUCTURES | | Descriptor: | Acetylcholine receptor subunit alpha | | Authors: | Lugovskoy, A.A, Maslennikov, I.V, Utkin, Y.N, Tsetlin, V.I, Cohen, J.B, Arseniev, A.S. | | Deposit date: | 1997-07-15 | | Release date: | 1998-01-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Spatial structure of the M3 transmembrane segment of the nicotinic acetylcholine receptor alpha subunit.
Eur.J.Biochem., 255, 1998
|
|
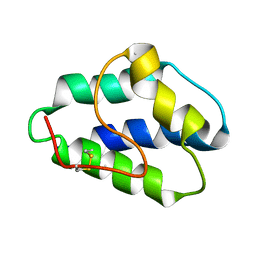
1LIP
 
 | |
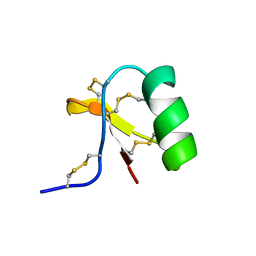
1C55
 
 | |
1C56
 
 | |
1KCN
 
 | | Structure of e109 Zeta Peptide, an Antagonist of the High-Affinity IgE Receptor | | Descriptor: | e109 zeta peptide | | Authors: | Nakamura, G.R, Reynolds, M.E, Chen, Y.M, Starovasnik, M.A, Lowman, H.B. | | Deposit date: | 2001-11-09 | | Release date: | 2002-03-06 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Stable "zeta" peptides that act as potent antagonists of the high-affinity IgE receptor.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KCO
 
 | | Structure of e131 Zeta Peptide, a Potent Antagonist of the High-Affinity IgE Receptor | | Descriptor: | e131 Zeta Peptide | | Authors: | Nakamura, G.R, Reynolds, M.E, Chen, Y.M, Starovasnik, M.A, Lowman, H.B. | | Deposit date: | 2001-11-09 | | Release date: | 2002-03-06 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Stable "zeta" peptides that act as potent antagonists of the high-affinity IgE receptor.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1IMW
 
 | | Peptide Antagonist of IGFBP-1 | | Descriptor: | IGFBP-1 antagonist | | Authors: | Lowman, H.B, Chen, Y.M, Skelton, N.J, Mortensen, D.L, Tomlinson, E.E, Sadick, M.D, Robinson, I.C, Clark, R.G. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
1NGL
 
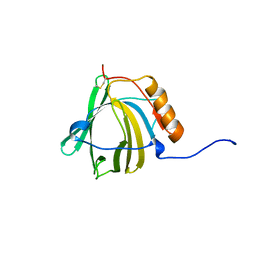 | | HUMAN NEUTROPHIL GELATINASE-ASSOCIATED LIPOCALIN (HNGAL), REGULARISED AVERAGE NMR STRUCTURE | | Descriptor: | PROTEIN (NGAL) | | Authors: | Coles, M, Diercks, T, Muehlenweg, B, Bartsch, S, Zoelzer, V, Tschesche, H, Kessler, H. | | Deposit date: | 1999-02-23 | | Release date: | 1999-05-26 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure and dynamics of human neutrophil gelatinase-associated lipocalin.
J.Mol.Biol., 289, 1999
|
|
2B7T
 
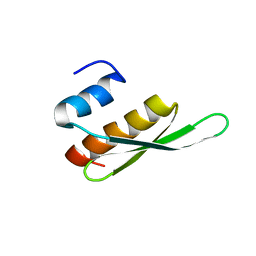 | | Structure of ADAR2 dsRBM1 | | Descriptor: | Double-stranded RNA-specific editase 1 | | Authors: | Stefl, R, Xu, M, Skrisovska, L, Emeson, R.B, Allain, F.H.-T. | | Deposit date: | 2005-10-05 | | Release date: | 2006-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and specific RNA binding of ADAR2 double-stranded RNA binding motifs.
Structure, 14, 2006
|
|
2B7V
 
 | | Structure of ADAR2 dsRBM2 | | Descriptor: | Double-stranded RNA-specific editase 1 | | Authors: | Stefl, R, Xu, M, Skrisovska, L, Emeson, R.B, Allain, F.H.-T. | | Deposit date: | 2005-10-05 | | Release date: | 2006-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and specific RNA binding of ADAR2 double-stranded RNA binding motifs.
Structure, 14, 2006
|
|
2BZ2
 
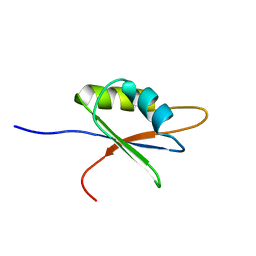 | | Solution structure of NELF E RRM | | Descriptor: | NEGATIVE ELONGATION FACTOR E | | Authors: | Schweimer, K, Rao, J.N, Neumann, L, Rosch, P, Wohrl, B.M. | | Deposit date: | 2005-08-10 | | Release date: | 2006-08-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural studies on the RNA-recognition motif of NELF E, a cellular negative transcription elongation factor involved in the regulation of HIV transcription.
Biochem. J., 400, 2006
|
|
3VDH
 
 | | Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14 | | Descriptor: | B-1,4-endoglucanase, CHLORIDE ION | | Authors: | Stogios, P.J, Evdokimova, E, Egorova, O, Yim, V, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-01-05 | | Release date: | 2012-01-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-Function Analysis of a Mixed-linkage beta-Glucanase/Xyloglucanase from the Key Ruminal Bacteroidetes Prevotella bryantii B14.
J.Biol.Chem., 291, 2016
|
|
8QCQ
 
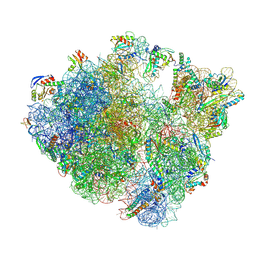 | | B. subtilis ApdA-stalled ribosomal complex | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Morici, M, Wilson, D.N. | | Deposit date: | 2023-08-28 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | RAPP-containing arrest peptides induce translational stalling by short circuiting the ribosomal peptidyltransferase activity.
Nat Commun, 15, 2024
|
|
8AS9
 
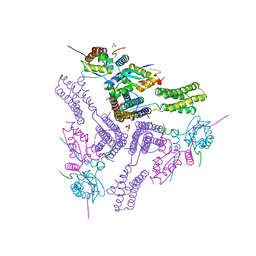 | | Crystal structure of the talin-KANK1 complex | | Descriptor: | B-cell lymphoma 6 protein, GLYCEROL, KN-motif NCoR1 BBD fusion,Nuclear receptor corepressor 1, ... | | Authors: | Zacharchenko, T. | | Deposit date: | 2022-08-18 | | Release date: | 2023-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The structural basis of the talin-KANK1 interaction that coordinates the actin and microtubule cytoskeletons at focal adhesions.
Open Biology, 13, 2023
|
|
6UH4
 
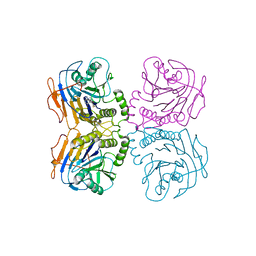 | | B. theta Bile Salt Hydrolase with covalent inhibitor | | Descriptor: | (5R,6R)-6-[(1S,2R,4aS,4bS,7R,8aS,10R,10aS)-7,10-dihydroxy-1,2,4b-trimethyltetradecahydrophenanthren-2-yl]-5-methylheptan-2-one, Choloylglycine hydrolase | | Authors: | Seegar, T.C.M. | | Deposit date: | 2019-09-26 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Development of a covalent inhibitor of gut bacterial bile salt hydrolases.
Nat.Chem.Biol., 16, 2020
|
|
5D9M
 
 | | Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, E280A mutant in complex with the xyloglucan tetradecasaccharide XXXGXXXG | | Descriptor: | B-1,4-endoglucanase, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Morar, M, Stogios, P.J, Xu, X, Cui, H, Di Leo, R, Yim, V, Savchenko, A. | | Deposit date: | 2015-08-18 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Function Analysis of a Mixed-linkage beta-Glucanase/Xyloglucanase from the Key Ruminal Bacteroidetes Prevotella bryantii B14.
J.Biol.Chem., 291, 2016
|
|
5D9N
 
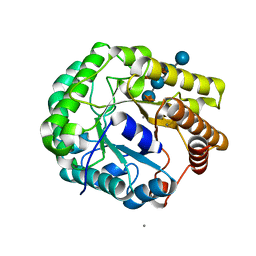 | | Crystal structure of PbGH5A, a glycoside hydrolase family 5 member from Prevotella bryantii B14, in complex with the xyloglucan heptasaccharide XXXG | | Descriptor: | B-1,4-endoglucanase, CALCIUM ION, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Morar, M, Stogios, P.J, Xu, X, Cui, H, Di Leo, R, Yim, V, Savchenko, A. | | Deposit date: | 2015-08-18 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-Function Analysis of a Mixed-linkage beta-Glucanase/Xyloglucanase from the Key Ruminal Bacteroidetes Prevotella bryantii B14.
J.Biol.Chem., 291, 2016
|
|
5D9O
 
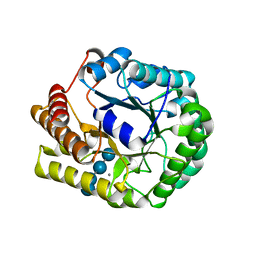 | | Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, E280A mutant in complex with cellotetraose | | Descriptor: | B-1,4-endoglucanase, CALCIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Morar, M, Stogios, P.J, Xu, X, Cui, H, Di Leo, R, Yim, V, Savchenko, A. | | Deposit date: | 2015-08-18 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Function Analysis of a Mixed-linkage beta-Glucanase/Xyloglucanase from the Key Ruminal Bacteroidetes Prevotella bryantii B14.
J.Biol.Chem., 291, 2016
|
|
