3MA3
 
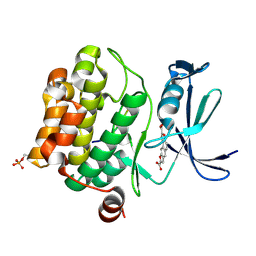 | | Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and a naphtho-difuran ligand | | Descriptor: | Pimtide, Proto-oncogene serine/threonine-protein kinase pim-1, naphtho[2,1-b:7,6-b']difuran-2,8-dicarboxylic acid | | Authors: | Filippakopoulos, P, Bullock, A, Fedorov, O, Vollmar, M, von Delft, F, Cochet, C, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-23 | | Release date: | 2010-04-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New potent dual inhibitors of CK2 and Pim kinases: discovery and structural insights.
Faseb J., 24, 2010
|
|
5PBJ
 
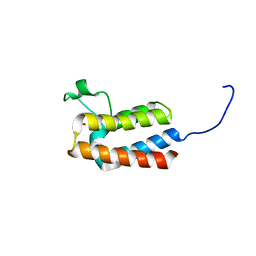 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 4) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PC0
 
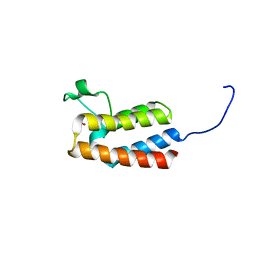 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 21) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
3V3X
 
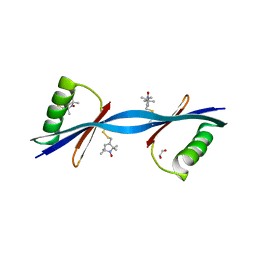 | | Nitroxide Spin Labels in Protein GB1: N8/K28 Double Mutant | | Descriptor: | ACETATE ION, GLYCEROL, Immunoglobulin G-binding protein G, ... | | Authors: | Cunningham, T.F, McGoff, M.S, Sengupta, I, Jaroniec, C.P, Horne, W.S, Saxena, S.K. | | Deposit date: | 2011-12-14 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structure of a protein spin-label in a solvent-exposed beta-sheet and comparison with DEER spectroscopy.
Biochemistry, 51, 2012
|
|
243L
 
 | | THE RESPONSE OF T4 LYSOZYME TO LARGE-TO-SMALL SUBSTITUTIONS WITHIN THE CORE AND ITS RELATION TO THE HYDROPHOBIC EFFECT | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, T4 LYSOZYME | | Authors: | Xu, J, Baase, W.A, Baldwin, E, Matthews, B.W. | | Deposit date: | 1997-10-22 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect.
Protein Sci., 7, 1998
|
|
5U4F
 
 | | Wild-type Transthyretin in complex with 1,1'-(1E)-(1,2-Ethenediyl)bis[2-chloro-4-boronic acid]benzene | | Descriptor: | Transthyretin, [(E)-ethene-1,2-diylbis(3-chloro-4,1-phenylene)]diboronic acid | | Authors: | Windsor, I.W, Smith, T.P, Raines, R.T, Forest, K.T. | | Deposit date: | 2016-12-03 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Stilbene Boronic Acids Form a Covalent Bond with Human Transthyretin and Inhibit Its Aggregation.
J. Med. Chem., 60, 2017
|
|
4OH4
 
 | | Crystal structure of BRI1 in complex with BKI1 | | Descriptor: | BRI1 kinase inhibitor 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Protein BRASSINOSTEROID INSENSITIVE 1 | | Authors: | Wang, J, Wang, J, Wu, J.W, Wang, Z.X. | | Deposit date: | 2014-01-17 | | Release date: | 2014-10-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into the negative regulation of BRI1 signaling by BRI1-interacting protein BKI1.
Cell Res., 24, 2014
|
|
4L7P
 
 | | Human artd3 (parp3) - catalytic domain in complex with inhibitor ME0395 | | Descriptor: | (2E)-N-[(2S)-1-hydroxy-3-phenylpropan-2-yl]-3-(4-oxo-1,4-dihydroquinazolin-2-yl)prop-2-enamide, Poly [ADP-ribose] polymerase 3 | | Authors: | Karlberg, T, Thorsell, A.G, Lindgren, A.E.G, Ekblad, T, Spjut, S, Andersson, C.D, Weigelt, J, Linusson, A, Elofsson, M, Schuler, H. | | Deposit date: | 2013-06-14 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Chemical Probes to Study ADP-Ribosylation: Synthesis and Biochemical Evaluation of Inhibitors of the Human ADP-Ribosyltransferase ARTD3/PARP3.
J.Med.Chem., 56, 2013
|
|
3V43
 
 | | Crystal structure of MOZ | | Descriptor: | ACETATE ION, Histone H3.1, Histone acetyltransferase KAT6A, ... | | Authors: | Qiu, Y, Li, F. | | Deposit date: | 2011-12-14 | | Release date: | 2012-06-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Combinatorial readout of unmodified H3R2 and acetylated H3K14 by the tandem PHD finger of MOZ reveals a regulatory mechanism for HOXA9 transcription
Genes Dev., 26, 2012
|
|
5PCN
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 44) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
3F3W
 
 | | Drug resistant cSrc kinase domain in complex with inhibitor RL45 (Type II) | | Descriptor: | 1-{4-[(6-aminoquinazolin-4-yl)amino]phenyl}-3-[3-tert-butyl-1-(3-methylphenyl)-1H-pyrazol-5-yl]urea, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Grutter, C, Kluter, S, Getlik, M, Rauh, D. | | Deposit date: | 2008-10-31 | | Release date: | 2009-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Hybrid compound design to overcome the gatekeeper T338M mutation in cSrc
J.Med.Chem., 52, 2009
|
|
5PD0
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 56) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PDG
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 72) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5J67
 
 | | Structure of Astrotactin-2, a conserved vertebrate-specific and perforin-like membrane protein involved in neuronal development | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Astrotactin-2, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Ni, T, Harlos, K, Gilbert, R.J.C. | | Deposit date: | 2016-04-04 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Structure of astrotactin-2: a conserved vertebrate-specific and perforin-like membrane protein involved in neuronal development.
Open Biology, 6, 2016
|
|
6M07
 
 | | Crystal structure of Lp-PLA2 in complex with a novel covalent inhibitor | | Descriptor: | (2S)-2-[(E)-3-[2-(diethylamino)ethyl-[[4-[4-(trifluoromethyl)-2-[2,2,2-tris(fluoranyl)ethoxy]phenyl]phenyl]methyl]amino]-1-oxidanyl-3-oxidanylidene-prop-1-enyl]pyrrolidine-1-carboxylic acid, Platelet-activating factor acetylhydrolase | | Authors: | Hu, H.C, Xu, Y.C. | | Deposit date: | 2020-02-20 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Identification of Highly Selective Lipoprotein-Associated Phospholipase A2 (Lp-PLA2) Inhibitors by a Covalent Fragment-Based Approach.
J.Med.Chem., 63, 2020
|
|
5V86
 
 | | Structure of DCN1 bound to NAcM-OPT | | Descriptor: | Lysozyme,DCN1-like protein 1, N-benzyl-N-(1-butylpiperidin-4-yl)-N'-(3,4-dichlorophenyl)urea | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.374 Å) | | Cite: | Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase.
Nat. Chem. Biol., 13, 2017
|
|
5PDW
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 89) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
4LCG
 
 | | Crystal structure of the Pseudomonas aeruginosa LPXC/LPC-050 complex | | Descriptor: | (betaS)-Nalpha-{4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]benzoyl}-N,beta-dihydroxy-L-tyrosinamide, NITRATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | Authors: | Lee, C.-J, Zhou, P. | | Deposit date: | 2013-06-21 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.568 Å) | | Cite: | Synthesis, Structure, and Antibiotic Activity of Aryl-Substituted LpxC Inhibitors.
J.Med.Chem., 56, 2013
|
|
5PEC
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 105) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
8AAF
 
 | | Yeast RQC complex in state G | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Tesina, P, Buschauer, R, Beckmann, R. | | Deposit date: | 2022-07-01 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular basis of eIF5A-dependent CAT tailing in eukaryotic ribosome-associated quality control.
Mol.Cell, 83, 2023
|
|
4ZFL
 
 | |
3ML5
 
 | | Crystal structure of the C183S/C217S mutant of human CA VII in complex with acetazolamide | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase 7, ZINC ION | | Authors: | Di Fiore, A, Truppo, E, Supuran, C.T, Alterio, V, Dathan, N, Bootorabi, F, Parkkila, S, Monti, S.M, De Simone, G. | | Deposit date: | 2010-04-16 | | Release date: | 2011-03-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the C183S/C217S mutant of human CA VII in complex with acetazolamide
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6B3P
 
 | | Crystal structure of CBMbc (family CBM26) from Eubacterium rectale Amy13K in Complex with Maltoheptaose | | Descriptor: | 1,2-ETHANEDIOL, Amy13K, FORMIC ACID, ... | | Authors: | Cockburn, D.W, Wawrzak, Z, Perez Medina, K, Koropatkin, N.M. | | Deposit date: | 2017-09-22 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Novel carbohydrate binding modules in the surface anchored alpha-amylase of Eubacterium rectale provide a molecular rationale for the range of starches used by this organism in the human gut.
Mol. Microbiol., 107, 2018
|
|
7JPZ
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI1 | | Descriptor: | (phenylmethyl) N-[(2S)-1-oxidanylidene-1-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-3-phenyl-propan-2-yl]carbamate, 3C-like proteinase | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
1LKT
 
 | | CRYSTAL STRUCTURE OF THE HEAD-BINDING DOMAIN OF PHAGE P22 TAILSPIKE PROTEIN | | Descriptor: | TAILSPIKE PROTEIN | | Authors: | Steinbacher, S. | | Deposit date: | 1997-10-17 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3 A, fully refined structure of the endorhamnosidase at 1.56 A resolution, and the molecular basis of O-antigen recognition and cleavage.
J.Mol.Biol., 267, 1997
|
|
