4RMZ
 
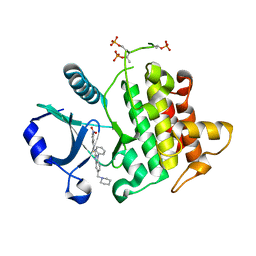 | | Crystal Structure of IRAK-4 | | Descriptor: | 3-nitro-N-[1-phenyl-5-(piperidin-1-ylmethyl)-1H-benzimidazol-2-yl]benzamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Johnstone, S, Sudom, A, Liu, J, Walker, N.P, Wang, Z. | | Deposit date: | 2014-10-22 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of IRAK-4 kinase in complex with inhibitors: a serine/threonine kinase with tyrosine as a gatekeeper
To be Published
|
|
5WBQ
 
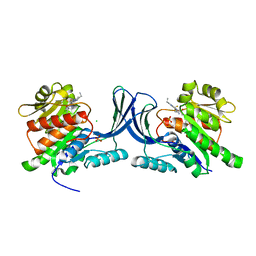 | | Structure of human Ketohexokinase complexed with hits from fragment screening | | Descriptor: | 2-ethyl-7-[(3S)-3-hydroxy-3-methylpyrrolidin-1-yl]-5-(trifluoromethyl)-1H-pyrrolo[3,2-b]pyridine-6-carbonitrile, CHLORIDE ION, Ketohexokinase, ... | | Authors: | Pandit, J. | | Deposit date: | 2017-06-29 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Fragment-Derived Small Molecules for in Vivo Inhibition of Ketohexokinase (KHK).
J. Med. Chem., 60, 2017
|
|
7ZZ8
 
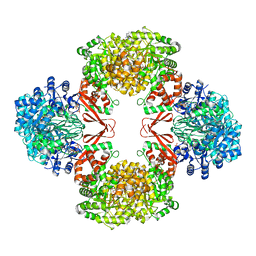 | | Cryo-EM structure of Lactococcus lactis pyruvate carboxylase with acetyl-CoA and cyclic di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, ACETYL COENZYME *A, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7V8Y
 
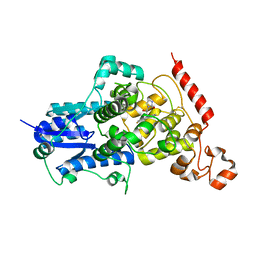 | | Crystal structure of mouse CRY2 in complex with SHP1703 compound | | Descriptor: | 1-[(2R)-3-[3,6-bis(fluoranyl)carbazol-9-yl]-2-oxidanyl-propyl]imidazolidin-2-one, Cryptochrome-2 | | Authors: | Miller, S.A, Hirota, T. | | Deposit date: | 2021-08-23 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CRY2 isoform selectivity of a circadian clock modulator with antiglioblastoma efficacy.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
9GBF
 
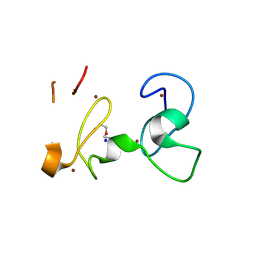 | | X-RAY structure of PHDvC5HCH tandem domain of NSD2 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, Histone-lysine N-methyltransferase NSD2, ... | | Authors: | Musco, G, Cocomazzi, P, Berardi, A, Knapp, S, Kramer, A. | | Deposit date: | 2024-07-31 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The C-terminal PHDVC5HCH tandem domain of NSD2 is a combinatorial reader of unmodified H3K4 and tri-methylated H3K27 that regulates transcription of cell adhesion genes in multiple myeloma.
Nucleic Acids Res., 53, 2025
|
|
9GKG
 
 | |
6DML
 
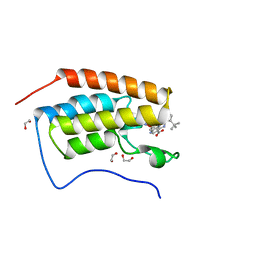 | | A multiconformer ligand model of 3,5 dimethylisoxaxole bound to the bromodomain of human BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 4-((2-(tert-butyl)phenyl)amino)-7-(3,5-dimethylisoxazol-4-yl)-6-methoxy-1,5-naphthyridine-3-carboxylic acid, Bromodomain-containing protein 4 | | Authors: | Hudson, B.M, van Zundert, G, Keedy, D.A, Fonseca, R, Heliou, A, Suresh, P, Borrelli, K, Day, T, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | qFit-ligand Reveals Widespread Conformational Heterogeneity of Drug-Like Molecules in X-Ray Electron Density Maps.
J. Med. Chem., 61, 2018
|
|
8RRT
 
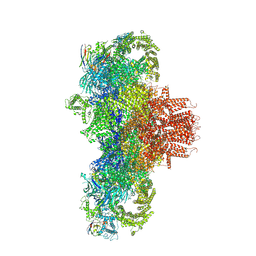 | |
8RRX
 
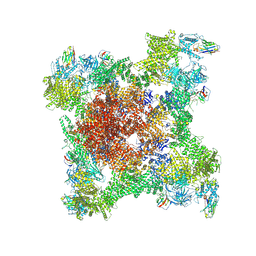 | | Structure of RyR1 reconstituted into lipid nanodisc in primed state in complex with Ca2+, ATP, caffeine and Nb9657 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, ... | | Authors: | Li, C, Efremov, R.G. | | Deposit date: | 2024-01-23 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Rapid small-scale nanobody-assisted purification of ryanodine receptors for cryo-EM.
J.Biol.Chem., 300, 2024
|
|
4Z02
 
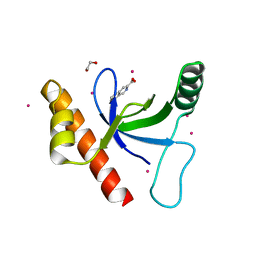 | | Crystal structure of BRD1 in complex with Isoquinoline-3-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, UNKNOWN ATOM OR ION, ... | | Authors: | DONG, A, IQBAL, A, WALKER, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-25 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of BRD1 incomplex with Isoquinoline-3-carboxylic acid
to be published
|
|
8RRW
 
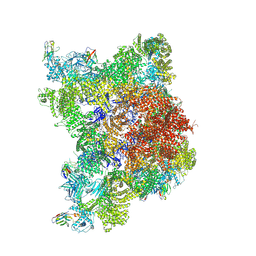 | |
8RS0
 
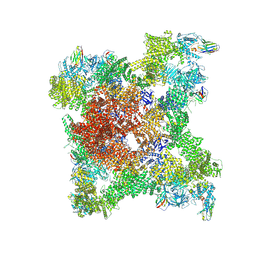 | |
8RRU
 
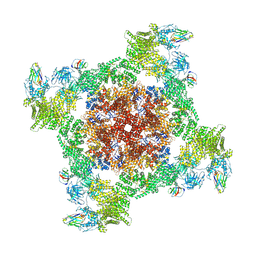 | |
9G67
 
 | | Crystal structure of Rhizobium etli L-asparaginase ReAV K138H mutant in complex with L-Asn | | Descriptor: | 1,2-ETHANEDIOL, ASPARAGINE, CADMIUM ION, ... | | Authors: | Pokrywka, K, Grzechowiak, M, Sliwiak, J, Worsztynowicz, P, Loch, J.I, Ruszkowski, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2024-07-18 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Controlling enzyme activity by mutagenesis and metal exchange to obtain crystal structures of stable substrate complexes of Class 3 l-asparaginase.
Febs J., 292, 2025
|
|
7T8V
 
 | | Co-crystal structure of Chaetomium glucosidase I with EB-0159 | | Descriptor: | (1S,2S,3R,4S,5S)-1-(hydroxymethyl)-5-[(6-{[2-nitro-4-(1H-1,2,3-triazol-1-yl)phenyl]amino}hexyl)amino]cyclohexane-1,2,3,4-tetrol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-12-17 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of Endoplasmic Reticulum alpha-Glucosidase I from a Thermophilic Fungus as a Platform for Structure-Guided Antiviral Drug Design.
Biochemistry, 61, 2022
|
|
8BWI
 
 | | Crystal structure of human Twisted gastrulation protein homolog 1 (TWSG1), crystal form 2 | | Descriptor: | Twisted gastrulation protein homolog 1 | | Authors: | Malinauskas, T, Rudolf, A.F, Moore, G, Eggington, H, Belnoue-Davis, H, El Omari, K, Woolley, R.E, Griffiths, S.C, Duman, R, Wagner, A, Leedham, S.J, Baldock, C, Ashe, H, Siebold, C. | | Deposit date: | 2022-12-06 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Molecular mechanism of BMP signal control by Twisted gastrulation.
Nat Commun, 15, 2024
|
|
4TU9
 
 | | STRUCTURE OF U2AF65 VARIANT WITH BRU5G6 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA (5'-D(*UP*UP*UP*UP*(BRU)P*DG*U)-3'), GLYCEROL, ... | | Authors: | Jenkins, J.L, McLaughlin, K.J, Agrawal, A.A, Kielkopf, C.L. | | Deposit date: | 2014-06-24 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Structure-guided U2AF65 variant improves recognition and splicing of a defective pre-mRNA.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7ZAZ
 
 | | macrocyclase OphP with ZPP | | Descriptor: | 1,2-ETHANEDIOL, BICARBONATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2022-03-23 | | Release date: | 2022-07-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the enzymatic macrocyclization of multiply backbone N-methylated peptides
Biorxiv, 2022
|
|
9B96
 
 | | Crystal structure of the human PAD2 protein bound to inhibitor | | Descriptor: | 1-({(2P)-1-{(1R)-1-(2-bromophenyl)-3-[5-(methanesulfonamido)-2-methylanilino]-3-oxopropyl}-2-[3-(4-chlorophenoxy)phenyl]-1H-1,3-benzimidazol-6-yl}methyl)-N-methyl-D-prolinamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Byrnes, L.J, Vajdos, F. | | Deposit date: | 2024-04-01 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery, Characterization, and Structure of a Cell Active PAD2 Inhibitor Acting through a Novel Allosteric Mechanism.
Acs Chem.Biol., 19, 2024
|
|
5YH5
 
 | | The crystal structure of Staphylococcus aureus CntA in apo form | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Nickel ABC transporter substrate-binding protein | | Authors: | Song, L, Ji, Q. | | Deposit date: | 2017-09-27 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanistic insights into staphylopine-mediated metal acquisition
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8SIW
 
 | | Structure of Compound 5 bound to the CHK1 10-point mutant | | Descriptor: | (1S,2S)-N-(7-chloro-6-{1-[(3R,4R)-4-hydroxy-3-methyloxolan-3-yl]piperidin-4-yl}isoquinolin-3-yl)-2-(1-methyl-1H-pyrazol-4-yl)cyclopropane-1-carboxamide, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2023-04-17 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.877 Å) | | Cite: | Discovery of MK-1468: A Potent, Kinome-Selective, Brain-Penetrant Amidoisoquinoline LRRK2 Inhibitor for the Potential Treatment of Parkinson's Disease.
J.Med.Chem., 66, 2023
|
|
6DBB
 
 | |
9BHI
 
 | | Crystal structure of the MerTK kinase domain with SA4488 | | Descriptor: | (5P)-2-amino-5-(1-methyl-1H-pyrazol-4-yl)-N-{(1R,2S)-2-[(4'-{2-[4-(2-oxoethyl)piperazin-1-yl]propan-2-yl}[1,1'-biphenyl]-4-yl)methoxy]cyclopentyl}pyridine-3-carboxamide, CHLORIDE ION, Mer tyrosine kinase domain | | Authors: | Jakob, C.G, Qui, W, Jain, R. | | Deposit date: | 2024-04-20 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of A-910, a Highly Potent and Orally Bioavailable Dual MerTK/Axl-Selective Tyrosine Kinase Inhibitor.
J.Med.Chem., 67, 2024
|
|
6QWV
 
 | | SARM1 SAM1-2 domains | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sporny, M, Isupov, N.M, Opatowsky, Y. | | Deposit date: | 2019-03-06 | | Release date: | 2019-07-03 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural Evidence for an Octameric Ring Arrangement of SARM1.
J.Mol.Biol., 431, 2019
|
|
4RNH
 
 | | PaMorA tandem diguanylate cyclase - phosphodiesterase, c-di-GMP complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, Motility regulator | | Authors: | Phippen, C.W, Tews, I. | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Formation and dimerization of the phosphodiesterase active site of the Pseudomonas aeruginosa MorA, a bi-functional c-di-GMP regulator.
Febs Lett., 588, 2014
|
|
