6D6N
 
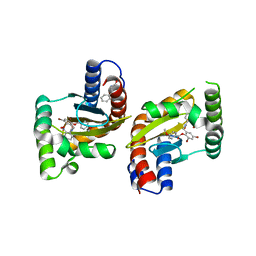 | | The structure of ligand binding domain of LasR in complex with TP-1 homolog, compound 16 | | Descriptor: | 2,4-dibromo-6-{[(2-nitrobenzene-1-carbonyl)amino]methyl}phenyl 4-methoxybenzoate, PHENYLALANINE, Transcriptional activator protein LasR | | Authors: | Dong, S.H, Nair, S.K. | | Deposit date: | 2018-04-21 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural and Biochemical Studies of Non-native Agonists of the LasR Quorum-Sensing Receptor Reveal an L3 Loop "Out" Conformation for LasR.
Cell Chem Biol, 25, 2018
|
|
6A0Q
 
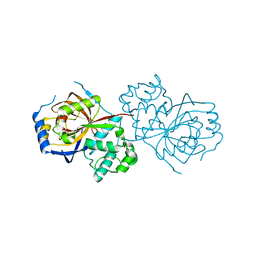 | | The crystal structure of Lpg2622_E64 complex | | Descriptor: | Lpg2622, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE | | Authors: | Gong, X, Ge, H. | | Deposit date: | 2018-06-06 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of the hypothetical protein Lpg2622, a new member of the C1 family peptidases from Legionella pneumophila
FEBS Lett., 592, 2018
|
|
5IIS
 
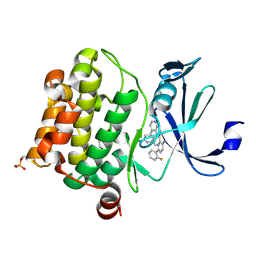 | | Design, synthesis and structure activity relationship of potent pan-PIM kinase inhibitors derived from the pyridyl-amide scaffold | | Descriptor: | 3-amino-N-(2'-amino-6'-methyl[4,4'-bipyridin]-3-yl)-6-(2-fluorophenyl)pyridine-2-carboxamide, DI(HYDROXYETHYL)ETHER, Serine/threonine-protein kinase pim-1 | | Authors: | Bellamacina, C, Bussiere, D, Burger, M. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, synthesis and structure activity relationship of potent pan-PIM kinase inhibitors derived from the pyridyl carboxamide scaffold.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
4QMN
 
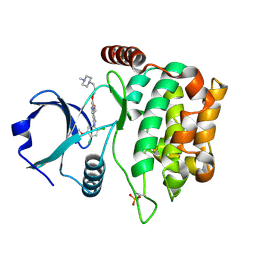 | | MST3 in complex with BOSUTINIB | | Descriptor: | 4-[(2,4-dichloro-5-methoxyphenyl)amino]-6-methoxy-7-[3-(4-methylpiperazin-1-yl)propoxy]quinoline-3-carbonitrile, Serine/threonine-protein kinase 24 | | Authors: | Olesen, S.H, Watts, C, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2014-06-16 | | Release date: | 2015-07-01 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Discovery of Diverse Small-Molecule Inhibitors of Mammalian Sterile20-like Kinase 3 (MST3).
Chemmedchem, 11, 2016
|
|
5LKJ
 
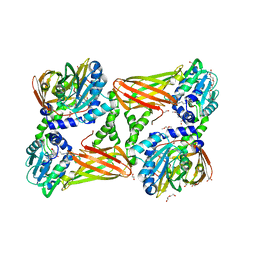 | | Crystal structure of mouse CARM1 in complex with ligand SA684 | | Descriptor: | (2~{S})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(2-carbamimidamidoethyl)amino]-2-azanyl-butanoic acid, 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-07-22 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with ligands
To Be Published
|
|
5BJY
 
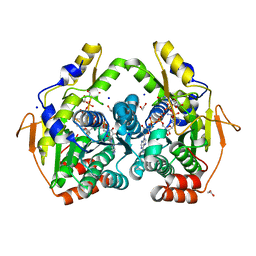 | | x-ray structure of the PglF 4,5-dehydratase from campylobacter jejuni, variant M405Y, in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Riegert, A.S, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-09-12 | | Release date: | 2017-11-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Investigation of PglF from Campylobacter jejuni Reveals a New Mechanism for a Member of the Short Chain Dehydrogenase/Reductase Superfamily.
Biochemistry, 56, 2017
|
|
4PY5
 
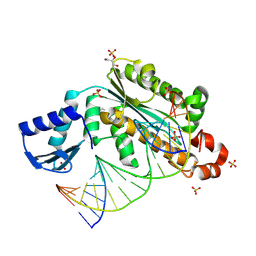 | | Thermovibrio ammonificans RNase H3 in complex with 19-mer RNA/DNA | | Descriptor: | 5'-D(*GP*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*CP*GP*CP*AP*CP*TP*C)-3', 5'-R(*GP*AP*GP*UP*GP*CP*GP*AP*CP*AP*CP*CP*UP*GP*AP*UP*UP*CP*C)-3', GLYCEROL, ... | | Authors: | Figiel, M, Nowotny, M. | | Deposit date: | 2014-03-26 | | Release date: | 2014-07-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of RNase H3-substrate complex reveals parallel evolution of RNA/DNA hybrid recognition.
Nucleic Acids Res., 42, 2014
|
|
8WM0
 
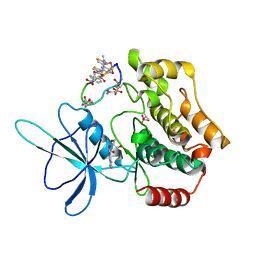 | | Crystal structure of TNIK-thiopeptide wTP3 complex | | Descriptor: | ADENOSINE, THIOPEPTIDE wTP3, TRAF2 and NCK-interacting protein kinase | | Authors: | Hamada, K, Kobayashi, S, Vinogradov, A.A, Zhang, Y, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2023-10-01 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Compact Reprogrammed Genetic Code for De Novo Discovery of Proteolytically Stable Thiopeptides.
J.Am.Chem.Soc., 2024
|
|
8CTI
 
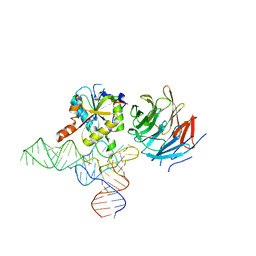 | | Cryo-EM structure of human METTL1-WDR4-tRNA(Val) complex | | Descriptor: | tRNA (guanine-N(7)-)-methyltransferase, tRNA (guanine-N(7)-)-methyltransferase non-catalytic subunit WDR4, tRNA-Val-TAC-2-1 | | Authors: | Li, J, Wang, L, Fontana, P, Hunkeler, M, Roy-Burman, S.S, Wu, H, Fischer, E.S, Gregory, R.I. | | Deposit date: | 2022-05-14 | | Release date: | 2022-12-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of regulated m 7 G tRNA modification by METTL1-WDR4.
Nature, 613, 2023
|
|
6DCC
 
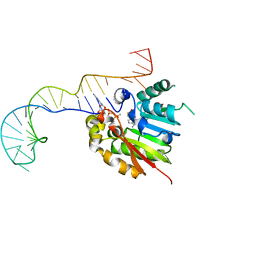 | | Structure of methylphosphate capping enzyme methyltransferase domain in complex with 5' end of 7SK RNA | | Descriptor: | 7SK snRNA methylphosphate capping enzyme, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Yang, Y, Eichhorn, C, Cascio, D, Feigon, J. | | Deposit date: | 2018-05-04 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of 7SK RNA 5'-gamma-phosphate methylation and retention by MePCE.
Nat. Chem. Biol., 15, 2019
|
|
7P39
 
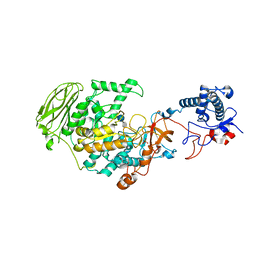 | | 4,6-alpha-glucanotransferase GtfB from Limosilactobacillus reuteri NCC 2613 complexed with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, CALCIUM ION, Dextransucrase, ... | | Authors: | Pijning, T, te Poele, E, Gangoiti, J, Boerner, T, Dijkhuizen, L. | | Deposit date: | 2021-07-07 | | Release date: | 2021-11-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights into Broad-Specificity Starch Modification from the Crystal Structure of Limosilactobacillus Reuteri NCC 2613 4,6-alpha-Glucanotransferase GtfB.
J.Agric.Food Chem., 69, 2021
|
|
4XU4
 
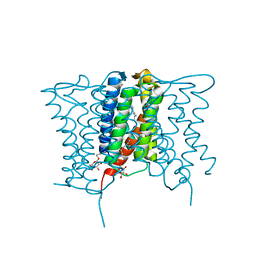 | | Crystal structure of a mycobacterial Insig homolog MvINS from Mycobacterium vanbaalenii at 1.9A resolution | | Descriptor: | DECYLAMINE-N,N-DIMETHYL-N-OXIDE, Uncharacterized protein, nonyl beta-D-glucopyranoside | | Authors: | Ren, R.B, Wu, J.P, Yan, C.Y, He, Y, Yan, N. | | Deposit date: | 2015-01-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | PROTEIN STRUCTURE. Crystal structure of a mycobacterial Insig homolog provides insight into how these sensors monitor sterol levels
Science, 349, 2015
|
|
5IGK
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with bromosporine (BSP) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, Bromosporine | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-28 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Promiscuous targeting of bromodomains by bromosporine identifies BET proteins as master regulators of primary transcription response in leukemia.
Sci Adv, 2, 2016
|
|
6FZ2
 
 | | Human N-myristoyltransferase (NMT1) with Myristoyl-CoA and inhibitor bound | | Descriptor: | GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, MAGNESIUM ION, ... | | Authors: | Kersten, F.C, Brenk, R, Jaenicke, E. | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | How To Design Selective Ligands for Highly Conserved Binding Sites: A Case Study UsingN-Myristoyltransferases as a Model System.
J.Med.Chem., 63, 2020
|
|
8AN8
 
 | | Crystal structure of wild-type c-MET bound by compound 7. | | Descriptor: | 3-[bis(fluoranyl)methyl]-~{N}-methyl-~{N}-[(1~{R})-8-methyl-5-(3-methyl-1~{H}-indazol-6-yl)-1,2,3,4-tetrahydronaphthalen-1-yl]pyridine-2-carboxamide, Hepatocyte growth factor receptor, SULFATE ION | | Authors: | Collie, G.W. | | Deposit date: | 2022-08-04 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Discovery of a selective c-MET inhibitor with a novel binding mode.
Bioorg.Med.Chem.Lett., 75, 2022
|
|
8DH1
 
 | | T7 RNA polymerase elongation complex with unnatural base dDs-PaTP pair | | Descriptor: | 1-{5-O-[(S)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-ribofuranosyl}-1H-pyrrole-2-carbaldehyde, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2022-06-24 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of transcription recognition of a hydrophobic unnatural base pair by T7 RNA polymerase.
Nat Commun, 14, 2023
|
|
7PD3
 
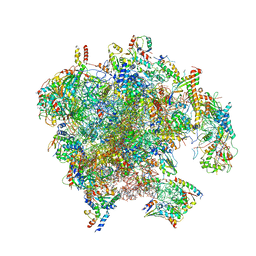 | | Structure of the human mitoribosomal large subunit in complex with NSUN4.MTERF4.GTPBP7 and MALSU1.L0R8F8.mt-ACP | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Chandrasekaran, V, Desai, N, Burton, N.O, Yang, H, Price, J, Miska, E.A, Ramakrishnan, V. | | Deposit date: | 2021-08-04 | | Release date: | 2021-11-17 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Visualizing formation of the active site in the mitochondrial ribosome.
Elife, 10, 2021
|
|
4XM6
 
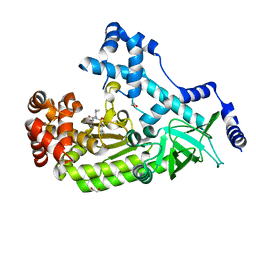 | | Anthrax toxin lethal factor with ligand-induced binding pocket | | Descriptor: | 1,2-ETHANEDIOL, Lethal factor, N~2~-[(4-fluoro-3-methylphenyl)sulfonyl]-N-hydroxy-N~2~-(2-methylpropyl)-D-valinamide, ... | | Authors: | Maize, K.M, De la Mora-Rey, T, Finzel, B.C. | | Deposit date: | 2015-01-14 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | Ligand-induced expansion of the S1' site in the anthrax toxin lethal factor.
Febs Lett., 589, 2015
|
|
1MJJ
 
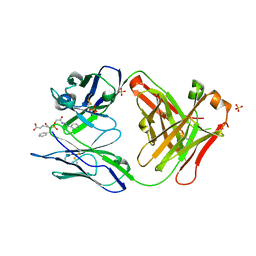 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF THE COMPLEX OF THE FAB FRAGMENT OF ESTEROLYTIC ANTIBODY MS6-12 AND A TRANSITION-STATE ANALOG | | Descriptor: | IMMUNOGLOBULIN MS6-12, N-{[2-({[1-(4-CARBOXYBUTANOYL)AMINO]-2-PHENYLETHYL}-HYDROXYPHOSPHINYL)OXY]ACETYL}-2-PHENYLETHYLAMINE, SULFATE ION | | Authors: | Ruzheinikov, S.N, Muranova, T.A, Sedelnikova, S.E, Partridge, L.J, Blackburn, G.M, Murray, I.A, Kakinuma, H, Takashi, N, Shimazaki, K, Sun, J, Nishi, Y, Rice, D.W. | | Deposit date: | 2002-08-28 | | Release date: | 2003-09-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution crystal structure of the Fab-fragments of a family of mouse catalytic antibodies with esterase activity
J.Mol.Biol., 332, 2003
|
|
5LMG
 
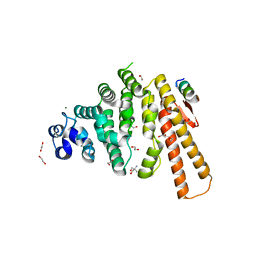 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator bound to Dcp2 HLM10 peptide (region 954-970) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA topoisomerase 2-associated protein PAT1, ... | | Authors: | Charenton, C, Gaudon-Plesse, C, Fourati, Z, Taverniti, V, Back, R, Kolesnikova, O, Seraphin, B, Graille, M. | | Deposit date: | 2016-07-30 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A unique surface on Pat1 C-terminal domain directly interacts with Dcp2 decapping enzyme and Xrn1 5'-3' mRNA exonuclease in yeast.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8W3A
 
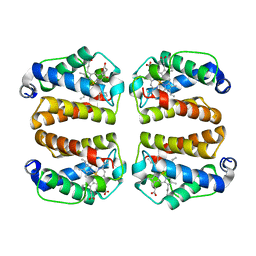 | | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin C51S C71S variant with trans heme D | | Descriptor: | Group 1 truncated hemoglobin, PROTOPORPHYRIN IX CONTAINING FE, {3-[(2R,5'R)-9',14'-diethenyl-5'-hydroxy-5',10',15',19'-tetramethyl-5-oxo-4,5-dihydro-3H-spiro[furan-2,4'-[21,22,23,24]tetraazapentacyclo[16.2.1.13,6.18,11.113,16]tetracosa[1,3(24),6,8,10,12,14,16(22),17,19]decaen]-20'-yl-kappa~4~N~21'~,N~22'~,N~23'~,N~24'~]propanoato}iron | | Authors: | Lecomte, J.T.J, Schlessman, J.L, Schultz, T.D, Siegler, M.A. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Heme d formation in a Shewanella benthica hemoglobin.
J.Inorg.Biochem., 259, 2024
|
|
8HWK
 
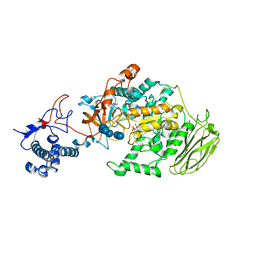 | | Limosilactobacillus reuteri N1 GtfB-maltohexaose | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Dong, J.J, Bai, Y.X. | | Deposit date: | 2022-12-30 | | Release date: | 2024-01-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights into the Structure-Function Relationship of GH70 GtfB alpha-Glucanotransferases from the Crystal Structure and Molecular Dynamic Simulation of a Newly Characterized Limosilactobacillus reuteri N1 GtfB Enzyme.
J.Agric.Food Chem., 72, 2024
|
|
8ARJ
 
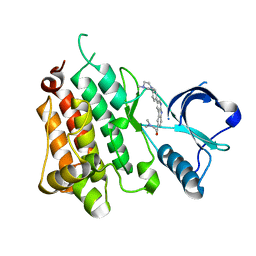 | | Anaplastic Lymphoma Kinase with a novel carboline inhibitor | | Descriptor: | 3-(dimethylamino)-1-[3-[4-(4-methylpiperazin-1-yl)phenyl]-9~{H}-pyrido[2,3-b]indol-6-yl]prop-2-en-1-one, ALK tyrosine kinase receptor | | Authors: | Mologni, L, Scapozza, L, Gambacorti-Passerini, C, Tardy, S, Goekjian, P, Robinson, C, Brown, D. | | Deposit date: | 2022-08-17 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.645 Å) | | Cite: | Discovery of Novel alpha-Carboline Inhibitors of the Anaplastic Lymphoma Kinase.
Acs Omega, 7, 2022
|
|
8W2L
 
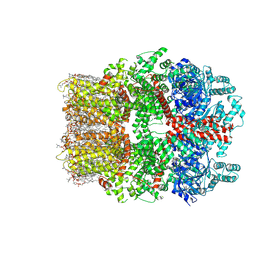 | | TRPM7 structure in complex with anticancer agent CCT128930 in closed state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 4-(4-chlorobenzyl)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-4-aminium, ... | | Authors: | Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2024-02-20 | | Release date: | 2024-04-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Structural basis of selective TRPM7 inhibition by the anticancer agent CCT128930.
Cell Rep, 43, 2024
|
|
5GIF
 
 | | Crystal Structure of Drosophila melanogaster E47D Dopamine N-Acetyltransferase in Binary Complex with Acetyl-CoA | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, ACETYL COENZYME *A, Dopamine N-acetyltransferase | | Authors: | Yang, Y.C, Wu, C.Y, Cheng, H.C, Lyu, P.C. | | Deposit date: | 2016-06-23 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Drosophila melanogaster E47D Dopamine N-Acetyltransferase in Binary Complex with Acetyl-CoA
To Be Published
|
|
