5MXV
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound GSK1107112A at 1.63A resolution | | Descriptor: | 3-chloranyl-~{N}-(4,5-dihydro-1,3-thiazol-2-yl)-6-fluoranyl-1-benzothiophene-2-carboxamide, HTH-type transcriptional regulator EthR | | Authors: | Blaszczyk, M, Mendes, V, Mugumbate, G, Blundell, T.L. | | Deposit date: | 2017-01-24 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.626 Å) | | Cite: | Target Identification of Mycobacterium tuberculosis Phenotypic Hits Using a Concerted Chemogenomic, Biophysical, and Structural Approach.
Front Pharmacol, 8, 2017
|
|
8S1K
 
 | | Crystal Structure of human FABP4 in complex with 2-[1-(methoxymethyl)cyclopentyl]-6-pentyl-4-phenyl-3-(1H-tetrazol-5-yl)-5,6,7,8-tetrahydroquinoline | | Descriptor: | (6~{S})-2-[1-(methoxymethyl)cyclopentyl]-6-pentyl-4-phenyl-3-(1~{H}-1,2,3,4-tetrazol-5-yl)-5,6,7,8-tetrahydroquinoline, DIMETHYL SULFOXIDE, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst-Sander, U, Rudolph, M.G. | | Deposit date: | 2024-02-15 | | Release date: | 2024-03-06 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. I. Dynamics of FABP4 and ligand binding.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
8P9L
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB512 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-azanyl-5-phenyl-phenyl)-5-(6-methyl-7-oxidanylidene-1~{H}-pyrrolo[2,3-c]pyridin-4-yl)pyridine-2-carboxamide | | Authors: | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-06 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
9JXZ
 
 | | V gamma9 V delta2 TCR and CD3 complex | | Descriptor: | 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, T cell receptor delta variable 2,T cell receptor delta constant, ... | | Authors: | Wang, L, Li, J, Li, Z. | | Deposit date: | 2024-10-12 | | Release date: | 2025-10-22 | | Last modified: | 2025-11-05 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structures of the apo and agonist-crosslinked human gamma delta T-cell receptor
To Be Published
|
|
6GRW
 
 | | Glucuronoyl Esterase from Opitutus terrae (Au derivative) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Lo Leggio, L, Larsbrink, J, Meland Knudsen, R, Mazurkewich, S, Navarro Poulsen, J.C. | | Deposit date: | 2018-06-12 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biochemical and structural features of diverse bacterial glucuronoyl esterases facilitating recalcitrant biomass conversion.
Biotechnol Biofuels, 11, 2018
|
|
8P9J
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB500 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-aminophenyl)-5-(6-methyl-7-oxidanylidene-1~{H}-pyrrolo[2,3-c]pyridin-4-yl)pyridine-2-carboxamide | | Authors: | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-06 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
8ANK
 
 | |
9AYL
 
 | | Cryo-EM structure of human Cav3.2 with ACT-709478 | | Descriptor: | 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2024-03-08 | | Release date: | 2024-04-24 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for human Ca v 3.2 inhibition by selective antagonists.
Cell Res., 34, 2024
|
|
8EDQ
 
 | | E. coli pyruvate kinase (PykF) I264F | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Pyruvate kinase, SULFATE ION | | Authors: | Donovan, K.A, Coombes, D, Dobson, R.C.J, Cooper, T.F. | | Deposit date: | 2022-09-05 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Beneficial mutations occurring in E. coli pyruvate kinase afford new allosteric mechanisms leading to faster resumption of growth
To Be Published
|
|
7RWI
 
 | | Mycobacterium tuberculosis RNA polymerase sigma L holoenzyme open promoter complex containing TNP-2198 | | Descriptor: | (3aM,9S,10bP,14S,15R,16S,17R,18R,19R,20S,21S,25R)-6,18,20-trihydroxy-14-methoxy-7,9,15,17,19,21,25-heptamethyl-1'-[2-(2-methyl-5-nitro-1H-imidazol-1-yl)ethyl]-5,10,26-trioxo-3,5,9,10-tetrahydrospiro[9,4-(epoxypentadecanoimino)furo[2',3':7,8]naphtho[1,2-d]imidazole-2,4'-piperidin]-16-yl acetate, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Molodtsov, V, Ebright, R.H. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Design, Synthesis, and Characterization of TNP-2198, a Dual-Targeted Rifamycin-Nitroimidazole Conjugate with Potent Activity against Microaerophilic and Anaerobic Bacterial Pathogens.
J.Med.Chem., 65, 2022
|
|
9KAU
 
 | |
8DTC
 
 | |
8U0R
 
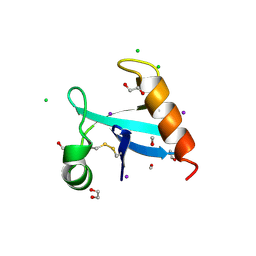 | | The crystal structure of protein A21, a component of the conserved poxvirus entry-fusion complex | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 2-(2-METHOXYETHOXY)ETHANOL, ... | | Authors: | Diesterbeck, U, Gittis, A.G, Garboczi, D.N, Moss, B. | | Deposit date: | 2023-08-29 | | Release date: | 2024-09-04 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The 2.3 angstrom Structure of A21, a Protein Component of the Conserved Poxvirus Entry-Fusion Complex.
J.Mol.Biol., 437, 2025
|
|
8DOJ
 
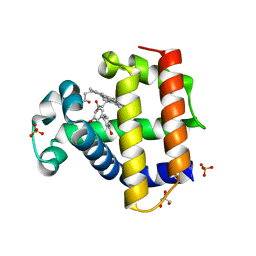 | | Dehaloperoxidase B in complex with 3,3'-Biphenol | | Descriptor: | (1P)-[1,1'-biphenyl]-3,3'-diol, Dehaloperoxidase B, GLYCEROL, ... | | Authors: | de Serrano, V.S, Yun, D, Ghiladi, R. | | Deposit date: | 2022-07-13 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Oxidation of bisphenol A (BPA) and related compounds by the multifunctional catalytic globin dehaloperoxidase.
J.Inorg.Biochem., 238, 2023
|
|
8TXL
 
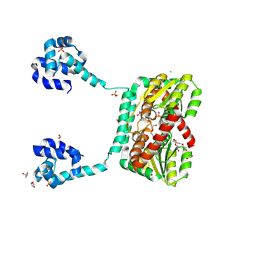 | |
9KBS
 
 | |
6PVS
 
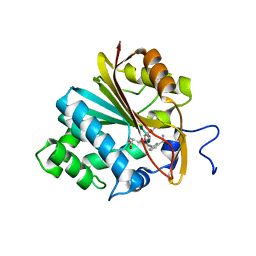 | | Structure of Nicotinamide N-Methyltransferase (NNMT) in complex with inhibitor LL320 | | Descriptor: | 9-(5-{[(3R)-3-amino-3-carboxypropyl][3-(3-carbamoylphenyl)prop-2-yn-1-yl]amino}-5-deoxy-alpha-D-lyxofuranosyl)-9H-purin-6-amine, NNMT protein | | Authors: | Noinaj, N, Huang, R, Chen, D, Yadav, R. | | Deposit date: | 2019-07-21 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.575 Å) | | Cite: | Novel Propargyl-Linked Bisubstrate Analogues as Tight-Binding Inhibitors for NicotinamideN-Methyltransferase.
J.Med.Chem., 62, 2019
|
|
7DBE
 
 | | Structure of a novel transaminase | | Descriptor: | 1,2-ETHANEDIOL, PYRIDOXAL-5'-PHOSPHATE, branched-chain amino acid aminotransferase | | Authors: | Li, F.L, Yu, H.M. | | Deposit date: | 2020-10-20 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and Structural Characterization of an (R)-Selective Transaminase in the Asymmetric Synthesis of Chiral Hydroxy Amines.
Adv.Synth.Catal., 2021
|
|
7UGE
 
 | | Bromodomain of CBP liganded with BMS-536924 | | Descriptor: | (3M)-4-{[(2S)-2-(3-chlorophenyl)-2-hydroxyethyl]amino}-3-[4-methyl-6-(morpholin-4-yl)-1H-benzimidazol-2-yl]pyridin-2(1H)-one, 1,2-ETHANEDIOL, Histone acetyltransferase | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2022-03-24 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Structural basis of CBP and EP300 interaction with kinase inhibitors
To Be Published
|
|
5MEK
 
 | | Sulphotransferase-18 from Arabidopsis thaliana in complex with 3'-phosphoadenosine 5'-phosphate (PAP) | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-3'-5'-DIPHOSPHATE, Cytosolic sulfotransferase 18, ... | | Authors: | Hirschmann, F, Krause, F, Baruch, P, Chizhov, I, Mueller, J.W, Manstein, D.J, Papenbrock, J, Fedorov, R. | | Deposit date: | 2016-11-15 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural and biochemical studies of sulphotransferase 18 from Arabidopsis thaliana explain its substrate specificity and reaction mechanism.
Sci Rep, 7, 2017
|
|
6YK2
 
 | | Structure of the AMPA receptor GluA2o ligand-binding domain (S1S2J) in complex with the compound (S)-1-[2'-Amino-2'-carboxyethyl]-5,7-dihydrothieno[3,4-d]pyrimidin- 2,4(1H,3H)-dione at resolution 1.60A | | Descriptor: | (2~{S})-2-azanyl-3-[2,4-bis(oxidanylidene)-5,7-dihydrothieno[3,4-d]pyrimidin-1-yl]propanoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2020-04-05 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.612 Å) | | Cite: | Ionotropic Glutamate Receptor GluA2 in Complex with Bicyclic Pyrimidinedione-Based Compounds: When Small Compound Modifications Have Distinct Effects on Binding Interactions.
Acs Chem Neurosci, 11, 2020
|
|
5GUL
 
 | | Crystal structure of Tris/PPix2/Mg2+ bound form of cyclolavandulyl diphosphate synthase (CLDS) from Streptomyces sp. CL190 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cyclolavandulyl diphosphate synthase, MAGNESIUM ION, ... | | Authors: | Tomita, T, Kobayashi, M, Nishiyama, M, Kuzuyama, T. | | Deposit date: | 2016-08-29 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure and Mechanism of the Monoterpene Cyclolavandulyl Diphosphate Synthase that Catalyzes Consecutive Condensation and Cyclization.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
8AU3
 
 | | c-MET Y1234E,Y1235E mutant in complex with Tepotinib | | Descriptor: | 3-[1-(3-{5-[(1-methylpiperidin-4-yl)methoxy]pyrimidin-2-yl}benzyl)-6-oxo-1,6-dihydropyridazin-3-yl]benzonitrile, DI(HYDROXYETHYL)ETHER, Hepatocyte growth factor receptor, ... | | Authors: | Graedler, U, Lammens, A. | | Deposit date: | 2022-08-25 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Biophysical and structural characterization of the impacts of MET phosphorylation on tepotinib binding.
J.Biol.Chem., 299, 2023
|
|
8AW1
 
 | | c-MET Y1235D mutant in complex with Tepotinib | | Descriptor: | 3-[1-(3-{5-[(1-methylpiperidin-4-yl)methoxy]pyrimidin-2-yl}benzyl)-6-oxo-1,6-dihydropyridazin-3-yl]benzonitrile, Hepatocyte growth factor receptor | | Authors: | Graedler, U, Lammens, A. | | Deposit date: | 2022-08-29 | | Release date: | 2023-09-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Biophysical and structural characterization of the impacts of MET phosphorylation on tepotinib binding.
J.Biol.Chem., 299, 2023
|
|
9B5P
 
 | |
