3FV1
 
 | | Crystal Structure of the human glutamate receptor, GluR5, ligand-binding core in complex with dysiherbaine in space group P1 | | Descriptor: | (2R,3aR,6S,7R,7aR)-2-[(2S)-2-amino-2-carboxyethyl]-6-hydroxy-7-(methylamino)hexahydro-2H-furo[3,2-b]pyran-2-carboxylic acid, GLYCEROL, Glutamate receptor, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2009-01-15 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
3FVK
 
 | | Crystal structure of the human glutamate receptor, GluR5, ligand-binding core in complex with 8-deoxy-neodysiherbaine A in space group P1 | | Descriptor: | (2R,3aR,6S,7aR)-2-[(2S)-2-amino-3-hydroxy-3-oxo-propyl]-6-hydroxy-3,3a,5,6,7,7a-hexahydrofuro[4,5-b]pyran-2-carboxylic acid, GLYCEROL, Glutamate receptor, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2009-01-15 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
3FVG
 
 | | Crystal structure of the human glutamate receptor, GluR5, ligand-binding core in complex with MSVIII-19 in space group P1 | | Descriptor: | (2R,3aR,7aR)-2-[(2S)-2-amino-3-hydroxy-3-oxo-propyl]-3,3a,5,6,7,7a-hexahydrofuro[4,5-b]pyran-2-carboxylic acid, GLYCEROL, Glutamate receptor, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2009-01-15 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
3FUZ
 
 | | Crystal structure of the human glutamate receptor, GluR5, ligand-binding core in complex with L-glutamate in space group P1 | | Descriptor: | GLUTAMIC ACID, Glutamate receptor, ionotropic kainate 1, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2009-01-15 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
3FVN
 
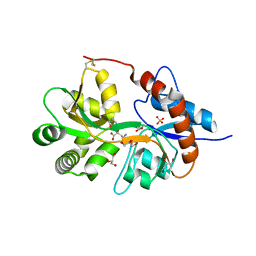 | | Crystal structure of the human glutamate receptor, GluR5, ligand-binding core in complex with 9-deoxy-neodysiherbaine A in space group P1 | | Descriptor: | (2R,3aR,7R,7aR)-2-[(2S)-2-amino-3-hydroxy-3-oxo-propyl]-7-hydroxy-3,3a,5,6,7,7a-hexahydrofuro[4,5-b]pyran-2-carboxylic acid, GLYCEROL, Glutamate receptor, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2009-01-16 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
3FV2
 
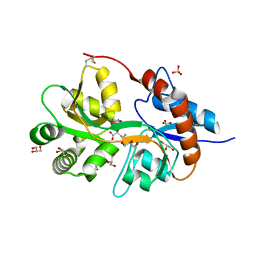 | | Crystal structure of the human glutamate receptor, GluR5, ligand-binding core in complex with neodysiherbaine A in space group P1 | | Descriptor: | (2R,3aR,6R,7R,7aR)-2-[(2S)-2-amino-2-carboxyethyl]-6,7-dihydroxyhexahydro-2H-furo[3,2-b]pyran-2-carboxylic acid, GLYCEROL, Glutamate receptor, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2009-01-15 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
2QS1
 
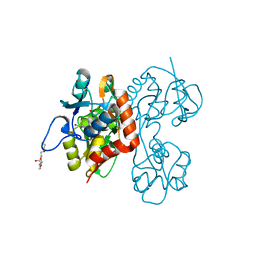 | | Crystal structure of the GluR5 ligand binding core dimer in complex with UBP315 at 1.80 Angstroms resolution | | Descriptor: | 3-({3-[(2S)-2-amino-2-carboxyethyl]-5-methyl-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl}methyl)-4,5-dibromothiophene-2-carboxylic acid, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Alushin, G.M, Jane, D.E, Mayer, M.L. | | Deposit date: | 2007-07-30 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists.
Neuropharmacology, 60, 2011
|
|
2QS4
 
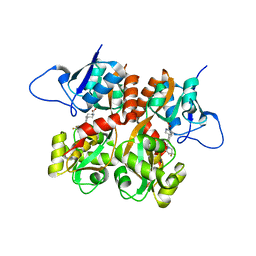 | | Crystal structure of the GluR5 ligand binding core dimer in complex with LY466195 at 1.58 Angstroms resolution | | Descriptor: | (3S,4aR,6S,8aR)-6-{[(2S)-2-carboxy-4,4-difluoropyrrolidin-1-yl]methyl}decahydroisoquinoline-3-carboxylic acid, AMMONIUM ION, GLYCEROL, ... | | Authors: | Alushin, G.M, Jane, D.E, Mayer, M.L. | | Deposit date: | 2007-07-30 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists.
Neuropharmacology, 60, 2011
|
|
2QS2
 
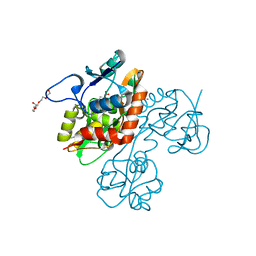 | | Crystal structure of the GluR5 ligand binding core dimer in complex with UBP318 at 1.80 Angstroms resolution | | Descriptor: | 3-({3-[(2S)-2-amino-2-carboxyethyl]-5-bromo-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl}methyl)thiophene-2-carboxylic acid, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Alushin, G.M, Jane, D.E, Mayer, M.L. | | Deposit date: | 2007-07-30 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists.
Neuropharmacology, 60, 2011
|
|
4P6Y
 
 | |
2D5K
 
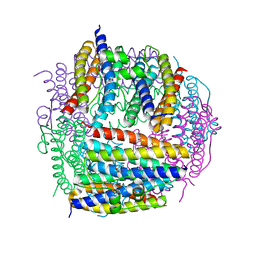 | | Crystal structure of Dps from Staphylococcus aureus | | Descriptor: | Dps family protein, GLYCEROL | | Authors: | Tanaka, Y, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2005-11-02 | | Release date: | 2006-10-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Nucleoid compaction by MrgA(Asp56Ala/Glu60Ala) does not contribute to staphylococcal cell survival against oxidative stress and phagocytic killing by macrophages
FEMS Microbiol. Lett., 360, 2014
|
|
3BV0
 
 | |
1Q55
 
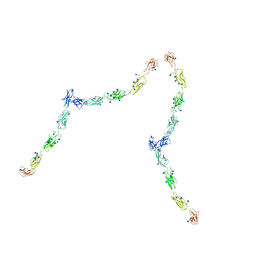 | | W-shaped trans interactions of cadherins model based on fitting C-cadherin (1L3W) to 3D map of desmosomes obtained by electron tomography | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | He, W, Cowin, P, Stokes, D.L. | | Deposit date: | 2003-08-06 | | Release date: | 2003-10-07 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (30 Å) | | Cite: | Untangling Desmosomal Knots with Electron Tomography
Science, 302, 2003
|
|
2KSQ
 
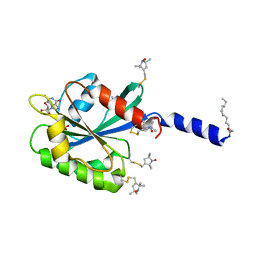 | | The myristoylated yeast ARF1 in a GTP and bicelle bound conformation | | Descriptor: | ADP-ribosylation factor 1, GUANOSINE-5'-TRIPHOSPHATE, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Liu, Y, Kahn, R, Prestegard, J. | | Deposit date: | 2010-01-12 | | Release date: | 2010-07-07 | | Last modified: | 2011-07-27 | | Method: | SOLUTION NMR | | Cite: | Dynamic structure of membrane-anchored Arf*GTP.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2MIA
 
 | |
2MI9
 
 | |
2LYI
 
 | |
2MEY
 
 | |
2M2G
 
 | | Solution structure of the antimicrobial peptide [Aba3,16]BTD-2 | | Descriptor: | [Aba3,16]BTD-2 | | Authors: | Conibear, A.C, Rosengren, K, Daly, N.L, Troiera Henriques, S, Craik, D.J. | | Deposit date: | 2012-12-20 | | Release date: | 2013-02-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The cyclic cystine ladder in theta-defensins is important for structure and stability, but not antibacterial activity.
J.Biol.Chem., 288, 2013
|
|
1WQ9
 
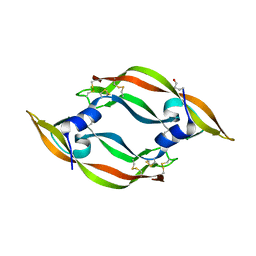 | | Crystal structure of VR-1, a VEGF-F from a snake venom | | Descriptor: | Vascular endothelial growth factor | | Authors: | Suto, K, Yamazaki, Y, Morita, T, Mizuno, H. | | Deposit date: | 2004-09-24 | | Release date: | 2004-12-07 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of novel vascular endothelial growth factors (VEGF) from snake venoms: insight into selective VEGF binding to kinase insert domain-containing receptor but not to fms-like tyrosine kinase-1.
J.Biol.Chem., 280, 2005
|
|
2LUF
 
 | |
2M2X
 
 | | Solution structure of the antimicrobial peptide [Aba3,5,7,12,14,16]BTD-2 | | Descriptor: | [Aba3,5,7,12,14,16]BTD-2 | | Authors: | Conibear, A.C, Rosengren, K, Daly, N.L, Troiera Henriques, S, Craik, D.J. | | Deposit date: | 2013-01-03 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | The cyclic cystine ladder in theta-defensins is important for structure and stability, but not antibacterial activity.
J.Biol.Chem., 288, 2013
|
|
1P8X
 
 | |
2M1P
 
 | | [Aba5,14]BTD-2 | | Descriptor: | [Aba5,14]BTD-2 | | Authors: | Conibear, A.C, Rosengren, K, Daly, N.L, Troiera Henriques, S, Craik, D.J. | | Deposit date: | 2012-12-04 | | Release date: | 2013-02-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The cyclic cystine ladder in theta-defensins is important for structure and stability, but not antibacterial activity.
J.Biol.Chem., 288, 2013
|
|
2UAG
 
 | | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PROTEIN (UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE), ... | | Authors: | Bertrand, J, Fanchon, E, Dideberg, O. | | Deposit date: | 1999-02-23 | | Release date: | 2000-02-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Determination of the MurD mechanism through crystallographic analysis of enzyme complexes.
J.Mol.Biol., 289, 1999
|
|
