8S0S
 
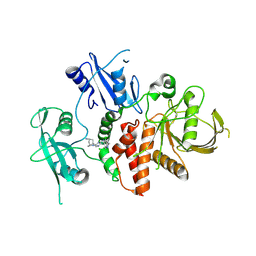 | | A fragment-based inhibitor of SHP2 | | Descriptor: | (1R,5S)-8-[7-(4-chloranyl-2-methyl-indazol-5-yl)-5H-pyrrolo[2,3-b]pyrazin-3-yl]-8-azabicyclo[3.2.1]octan-3-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Cleasby, A, Price, A. | | Deposit date: | 2024-02-14 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Fragment-Based Discovery of Allosteric Inhibitors of SH2 Domain-Containing Protein Tyrosine Phosphatase-2 (SHP2).
J.Med.Chem., 67, 2024
|
|
6NM4
 
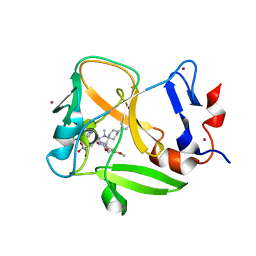 | | Crystal structure of SAM-bound PRDM9 in complex with MRK-740 inhibitor | | Descriptor: | 4-[3-(3,5-dimethoxyphenyl)-1,2,4-oxadiazol-5-yl]-1-methyl-9-(2-methylpyridin-4-yl)-1,4,9-triazaspiro[5.5]undecane, Histone-lysine N-methyltransferase PRDM9, S-ADENOSYLMETHIONINE, ... | | Authors: | Ivanochko, D, Halabelian, L, Fischer, C, Sanders, J.M, Kattar, S.D, Brown, P.J, Edwards, A.M, Bountra, C, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Discovery of a chemical probe for PRDM9.
Nat Commun, 10, 2019
|
|
6X8Q
 
 | |
6HK7
 
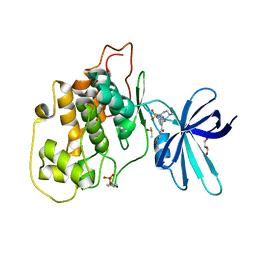 | | Crystal structure of GSK-3B in complex with pyrazine inhibitor C50 | | Descriptor: | 3-azanyl-~{N}-(2-methoxyethyl)-6-[4-(4-methylpiperazin-1-yl)sulfonylphenyl]pyrazine-2-carboxamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Piretti, V, Giabbai, B, Demitri, N, Di Martino, R, Tripathi, S.K, Gobbo, D, Decherchi, S, Storici, P, Girotto, S, Cavalli, A. | | Deposit date: | 2018-09-05 | | Release date: | 2019-07-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Investigating Drug-Target Residence Time in Kinases through Enhanced Sampling Simulations.
J Chem Theory Comput, 15, 2019
|
|
7AN5
 
 | | Enzyme of biosynthetic pathway | | Descriptor: | 3-[(1-Carboxyvinyl)oxy]benzoic acid, Chorismate dehydratase | | Authors: | Archna, A, Breithaupt, C, Stubbs, M.T. | | Deposit date: | 2020-10-11 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Mechanism of chorismate dehydratase MqnA, the first enzyme of the futalosine pathway, proceeds via substrate-assisted catalysis
J.Biol.Chem., 2022
|
|
5M46
 
 | | Alpha-amino epsilon-caprolactam racemase (ACLR) from Rhizobacterium freirei | | Descriptor: | 1,2-ETHANEDIOL, Aminotransferase class-III, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Frese, A, Sutton, P.W, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2016-10-18 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Snapshots of the Catalytic Cycle of the Industrial Enzyme alpha-Amino-epsilon-Caprolactam Racemase (ACLR) Observed Using X-ray Crystallography
Acs Catalysis, 7, 2017
|
|
5LLK
 
 | | Crystal structure of human alpha-dystroglycan | | Descriptor: | 1,2-ETHANEDIOL, Dystroglycan | | Authors: | Covaceuszach, S, Cassetta, A, Lamba, D, Brancaccio, A, Bozzi, M, Sciandra, F, Bigotti, M.G, Konarev, P.V. | | Deposit date: | 2016-07-27 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural flexibility of human alpha-dystroglycan.
FEBS Open Bio, 7, 2017
|
|
6HZ5
 
 | | Structure of McrBC without DNA binding domains (Class 1) | | Descriptor: | 5-methylcytosine-specific restriction enzyme B, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Itoh, Y, Nirwan, N, Saikrishnan, K, Amunts, A. | | Deposit date: | 2018-10-22 | | Release date: | 2019-07-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure-based mechanism for activation of the AAA+ GTPase McrB by the endonuclease McrC.
Nat Commun, 10, 2019
|
|
6NSQ
 
 | | Crystal structure of BRAF kinase domain bound to the inhibitor 2l | | Descriptor: | 5-[(4-amino-1-ethyl-1H-pyrazolo[3,4-d]pyrimidin-3-yl)ethynyl]-N-(4-chlorophenyl)-6-methylisoquinolin-1-amine, Serine/threonine-protein kinase B-raf | | Authors: | Maisonneuve, P, Kurinov, I, Assadieskandar, A, Yu, C, Zhang, C, Sicheri, F. | | Deposit date: | 2019-01-25 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Rigidification Dramatically Improves Inhibitor Selectivity for RAF Kinases.
Acs Med.Chem.Lett., 10, 2019
|
|
4Z7J
 
 | | Structure of Acetobacter aceti PurE-S57A | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, 5-O-phosphono-beta-D-ribofuranose, ... | | Authors: | Kappock, T.J, Sullivan, K.L. | | Deposit date: | 2015-04-07 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Mutation of the conserved serine in two PurE classes
To Be Published
|
|
8XF9
 
 | | High-resolution structure of the siderophore periplasmic binding protein FtsB mutant Y137A from Streptococcus pyogenes | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes.
Structure, 32, 2024
|
|
6XI9
 
 | | X-ray crystal structure of MqnE from Pedobacter heparinus in complex with aminofutalosine and methionine | | Descriptor: | 9-[7-(3-carboxyphenyl)-5,6-dideoxy-beta-D-ribo-heptodialdo-1,4-furanosyl]-9H-purin-6-amine, Aminodeoxyfutalosine synthase, CHLORIDE ION, ... | | Authors: | Grove, T.L, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2020-06-19 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Narrow-Spectrum Antibiotic Targeting of the Radical SAM Enzyme MqnE in Menaquinone Biosynthesis.
Biochemistry, 59, 2020
|
|
8EER
 
 | |
7BQK
 
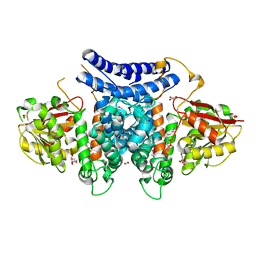 | | The structure of PdxI in complex with its substrate analogue | | Descriptor: | 1,2-ETHANEDIOL, 3-[(E,2S,4S)-2,4-dimethyloct-6-enoyl]-4-oxidanyl-1H-pyridin-2-one, GLYCEROL, ... | | Authors: | Cai, Y.J, Ohashi, M, Zhou, J.H, Tang, Y. | | Deposit date: | 2020-03-24 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | An enzymatic Alder-ene reaction.
Nature, 586, 2020
|
|
8XL1
 
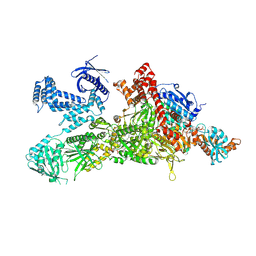 | | Core region of the human acetyl-CoA carboxylase 1 filament in complex with acetyl-CoA (ACC1-inact) | | Descriptor: | ACETYL COENZYME *A, Acetyl-CoA carboxylase 1 | | Authors: | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | Deposit date: | 2023-12-25 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Filament structures unveil the dynamic organization of human acetyl-CoA carboxylase.
Sci Adv, 10, 2024
|
|
8XFA
 
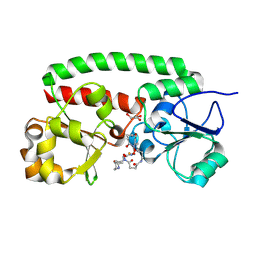 | | Structure of the siderophore periplasmic binding protein FtsB mutant Y137A from Streptococcus pyogenes with ferrioxamine E bound | | Descriptor: | (8E)-6,17,28-trihydroxy-1,6,12,17,23,28-hexaazacyclotritriacont-8-ene-2,5,13,16,24,27-hexone, FE (III) ION, GLYCEROL, ... | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes.
Structure, 32, 2024
|
|
8XN1
 
 | | Cryo-EM structure of human ZnT3, in the presence of zinc, determined in an inward-facing conformation | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, CHOLESTEROL HEMISUCCINATE, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Zhu, Z, Qu, Q, Long, Y, Zhou, Z. | | Deposit date: | 2023-12-28 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural insights into human zinc transporter ZnT1 mediated Zn 2+ efflux.
Embo Rep., 25, 2024
|
|
9GGU
 
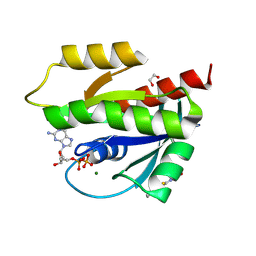 | | Human KRas4A (GDP) in complex with compound 9 | | Descriptor: | 1,2-ETHANEDIOL, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Schuettelkopf, A.W. | | Deposit date: | 2024-08-14 | | Release date: | 2025-04-16 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Reversible Small Molecule Multivariant Ras Inhibitors Display Tunable Affinity for the Active and Inactive Forms of Ras.
J.Med.Chem., 68, 2025
|
|
8XL0
 
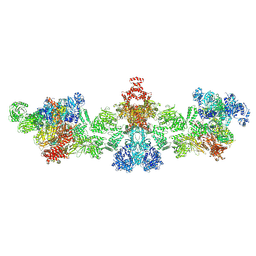 | | Citrate-induced filament of human acetyl-coenzyme A carboxylase 1 (ACC1-citrate) | | Descriptor: | Acetyl-CoA carboxylase 1, BIOTIN | | Authors: | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | Deposit date: | 2023-12-25 | | Release date: | 2024-10-23 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Filament structures unveil the dynamic organization of human acetyl-CoA carboxylase.
Sci Adv, 10, 2024
|
|
8JJP
 
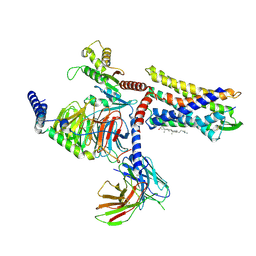 | | G protein-coupled receptor 1 | | Descriptor: | CHOLESTEROL, Chemerin-like receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, A, Liu, Y, Chen, G, Ye, F. | | Deposit date: | 2023-05-31 | | Release date: | 2024-08-21 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of G protein-coupled receptor GPR1 bound to full-length chemerin adipokine reveals a chemokine-like reverse binding mode.
Plos Biol., 22, 2024
|
|
7ZT9
 
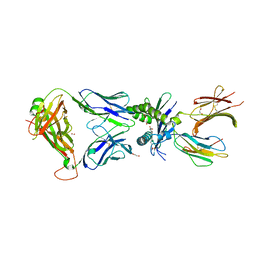 | | Structure of E8 TCR in complex in human MR1 bound to 4FBA | | Descriptor: | 1,2-ETHANEDIOL, 4-METHYLBENZOIC ACID, Beta-2-microglobulin, ... | | Authors: | Karuppiah, V, Srikannathasan, V, Robinson, R.A. | | Deposit date: | 2022-05-09 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Promiscuous recognition of MR1 drives self-reactive mucosal-associated invariant T cell responses.
J.Exp.Med., 220, 2023
|
|
8XKZ
 
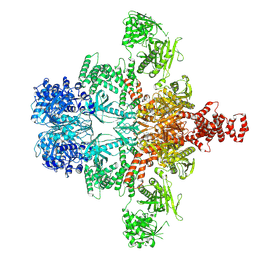 | | Core region of the citrate-induced human acetyl-CoA carboxylase 1 filament (ACC1-citrate) | | Descriptor: | Acetyl-CoA carboxylase 1, BIOTIN | | Authors: | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | Deposit date: | 2023-12-25 | | Release date: | 2024-10-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Filament structures unveil the dynamic organization of human acetyl-CoA carboxylase.
Sci Adv, 10, 2024
|
|
9B8P
 
 | | Synaptic Vesicle V-ATPase with synaptophysin and SidK, State 3, V1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase H+-transporting V1 subunit D, H(+)-transporting two-sector ATPase, ... | | Authors: | Coupland, E.M, Rubinstein, J.L. | | Deposit date: | 2024-03-31 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution electron cryomicroscopy of V-ATPase in native synaptic vesicles.
Science, 385, 2024
|
|
7BQ4
 
 | | X-ray structure of human PPARalpha ligand binding domain-eicosapentaenoic acid (EPA)-SRC1 coactivator peptide co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 5,8,11,14,17-EICOSAPENTAENOIC ACID, GLYCEROL, ... | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | Deposit date: | 2020-03-23 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6NJU
 
 | | Mouse endonuclease G mutant H97A bound to A-DNA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DNA (5'-D(CCGGCGCCGG)-3'), ... | | Authors: | Vander Zanden, C.M, Ho, E.N, Czarny, R.S, Robertson, A.B, Ho, P.S. | | Deposit date: | 2019-01-04 | | Release date: | 2020-01-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural adaptation of vertebrate endonuclease G for 5-hydroxymethylcytosine recognition and function.
Nucleic Acids Res., 48, 2020
|
|
