4NEL
 
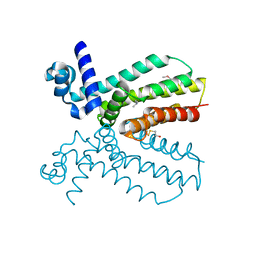 | | Crystal structure of a putative transcriptional regulator from Saccharomonospora viridis in complex with N,N-dimethylmethanamine | | Descriptor: | N,N-dimethylmethanamine, Transcriptional regulator | | Authors: | Halavaty, A.S, Filippova, E.V, Minasov, G, Kiryukhina, O, Shuvalova, L, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-10-29 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a putative transcriptional regulator from Saccharomonospora viridis in complex with N,N-dimethylmethanamine
To be Published
|
|
5VXN
 
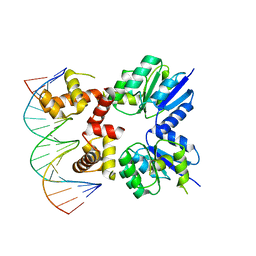 | | Structure of two RcsB dimers bound to two parallel DNAs. | | Descriptor: | DNA (5'-D(*GP*AP*TP*TP*TP*AP*GP*GP*AP*AP*AP*AP*AP*TP*CP*TP*TP*AP*GP*AP*TP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*GP*GP*AP*AP*AP*AP*AP*TP*CP*TP*TP*AP*GP*A)-3'), Transcriptional regulatory protein RcsB | | Authors: | Filippova, E.V, Minasov, G, Pshenychnyi, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-05-23 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.375 Å) | | Cite: | Structural Basis for DNA Recognition by the Two-Component Response Regulator RcsB.
MBio, 9, 2018
|
|
4MBR
 
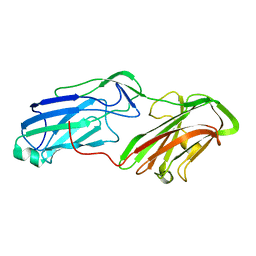 | | 3.65 Angstrom Crystal Structure of Serine-rich Repeat Protein (Srr2) from Streptococcus agalactiae | | Descriptor: | Serine-rich repeat protein 2 | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Seo, H.S, Seepersaud, R, Doran, K.S, Iverson, T.M, Sullam, P.M, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Characterization of Fibrinogen Binding by Glycoproteins Srr1 and Srr2 of Streptococcus agalactiae.
J.Biol.Chem., 288, 2013
|
|
2XMY
 
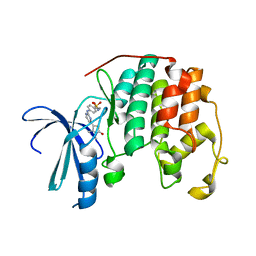 | | Discovery and Characterisation of 2-Anilino-4-(thiazol-5-yl) pyrimidine Transcriptional CDK Inhibitors as Anticancer Agents | | Descriptor: | 4-[4-(3,4-DIMETHYL-2-OXO-2,3-DIHYDRO-THIAZOL-5-YL)-PYRIMIDIN-2-YLAMINO]-N-(2-METHOXY-ETHYL)-BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Wang, S, Griffiths, G, Midgley, C.A, Barnett, A.L, Cooper, M, Grabarek, J, Ingram, L, Jackson, W, Kontopidis, G, McClue, S.J, McInnes, C, McLachlan, J, Meades, C, Mezna, M, Stuart, I, Thomas, M.P, Zheleva, D.I, Lane, D.P, Jackson, R.C, Glover, D.M, Blake, D.G, Fischer, P.M. | | Deposit date: | 2010-07-29 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Characterisation of 2-Anilino-4-(Thiazol-5-Yl)Pyrimidine Transcriptional Cdk Inhibitors as Anticancer Agents
Chem.Biol., 17, 2010
|
|
2UYW
 
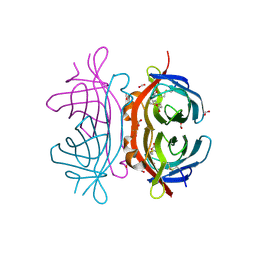 | | Crystal structure of Xenavidin | | Descriptor: | BIOTIN, FORMIC ACID, XENAVIDIN | | Authors: | Helppolainen, S.H, Maatta, J.A.E, Airenne, T.T, Johnson, M.S, Kulomaa, M.S, Nordlund, H.R. | | Deposit date: | 2007-04-20 | | Release date: | 2008-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Characteristics of Xenavidin, the First Frog Avidin from Xenopus Tropicalis.
Bmc Struct.Biol., 9, 2009
|
|
4ODI
 
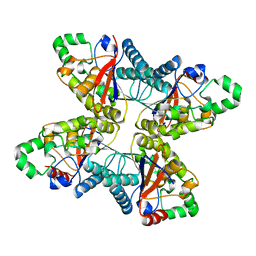 | | 2.6 Angstrom Crystal Structure of Putative Phosphoglycerate Mutase 1 from Toxoplasma gondii | | Descriptor: | Phosphoglycerate mutase PGMII, SODIUM ION | | Authors: | Minasov, G, Ruan, J, Ngo, H, Shuvalova, L, Dubrovska, I, Flores, K, Shanmugam, D, Roos, D, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-10 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
4CBE
 
 | | Crystal structure of complement factors H and FHL-1 binding protein BBH06 or CRASP-2 from Borrelia burgdorferi (Native) | | Descriptor: | COMPLEMENT REGULATOR-ACQUIRING SURFACE PROTEIN 2 (CRASP-2) | | Authors: | Brangulis, K, Petrovskis, I, Kazaks, A, Ranka, R, Tars, K. | | Deposit date: | 2013-10-13 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Characterization of Cspz, a Complement Regulator Factor H and Fhl-1 Binding Protein from Borrelia Burgdorferi.
FEBS J., 281, 2014
|
|
4D1W
 
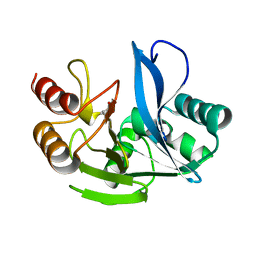 | | A H224Y mutant for VIM-7 from Pseudomonas aeruginosa | | Descriptor: | METALLO-B-LACTAMASE, ZINC ION | | Authors: | Leiros, H.-K.S, Skagseth, S, Edvardsen, K.S.W, Lorentzen, M.S, Bjerga, G.E.K, Leiros, I, Samuelsen, O. | | Deposit date: | 2014-05-05 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | His224 Alters the R2 Drug Binding Site and Phe218 Influences the Catalytic Efficiency in the Metallo-Beta-Lactamase Vim-7.
Antimicrob.Agents Chemother., 58, 2014
|
|
4CWA
 
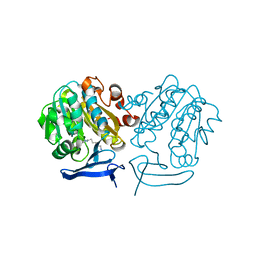 | | Structure of Plasmodium Falciparum Spermidine Synthase in Complex with 1H-Benzimidazole-2-pentanamine | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 5-(1H-benzimidazol-2-yl)pentan-1-amine, SPERMIDINE SYNTHASE | | Authors: | Sprenger, J, Halander, J.C, Svensson, B, Al-Karadaghi, S, Persson, L. | | Deposit date: | 2014-04-01 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Three-Dimensional Structures of Plasmodium Falciparum Spermidine Synthase with Bound Inhibitors Suggest New Strategies for Drug Design.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4CXM
 
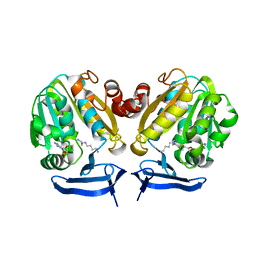 | | Crystal Structure of Plasmodium Falciparum Spermidine Synthase in Complex with METHYLTHIOADENOSIN AND SPERMIDINE after catalysis in crystal | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, SPERMIDINE, SPERMIDINE SYNTHASE | | Authors: | Sprenger, J, Svensson, B, Al-Karadaghi, S, Persson, L. | | Deposit date: | 2014-04-07 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Three-Dimensional Structures of Plasmodium Falciparum Spermidine Synthase with Bound Inhibitors Suggest New Strategies for Drug Design.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4D7U
 
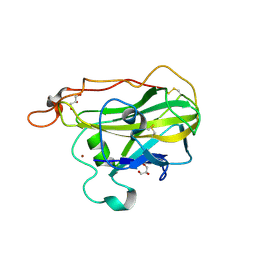 | | The structure of the catalytic domain of NcLPMO9C from the filamentous fungus Neurospora crassa | | Descriptor: | COPPER (II) ION, ENDOGLUCANASE II, GLYCEROL | | Authors: | Borisova, A.S, Isaksen, T, Mathiesen, G, Sorlie, M, Sandgren, M, Eijsink, V.G.H, Dimarogona, M. | | Deposit date: | 2014-11-27 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural and Functional Characterization of a Lytic Polysaccharide Monooxygenase with Broad Substrate Specificity
J.Biol.Chem., 290, 2015
|
|
4CNS
 
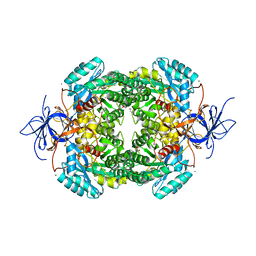 | | Crystal structure of truncated human CRMP-4 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ponnusamy, R, Lohkamp, B. | | Deposit date: | 2014-01-24 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Human Crmp-4: Correction of Intensities for Lattice-Translocation Disorder
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4KWA
 
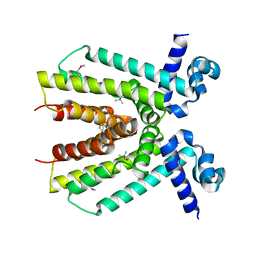 | | Crystal structure of a putative transcriptional regulator from Saccharomonospora viridis in complex with choline | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, CHOLINE ION, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-23 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a putative transcriptional regulator from Saccharomonospora viridis in complex with choline
To be Published
|
|
4CNU
 
 | |
4CML
 
 | | Crystal Structure of INPP5B in complex with Phosphatidylinositol 3,4- bisphosphate | | Descriptor: | 1,2-dioctanoyl phosphatidyl epi-inositol (3,4)-bisphosphate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tresaugues, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Edwards, A.M, Ekblad, T, Flodin, S, Graslund, S, Karlberg, T, Moche, M, Nyman, T, Schuler, H, Silvander, C, Thorsell, A.G, Weigelt, J, Welin, M, Nordlund, P. | | Deposit date: | 2014-01-16 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Phosphoinositide Substrate Recognition, Catalysis, and Membrane Interactions in Human Inositol Polyphosphate 5-Phosphatases.
Structure, 22, 2014
|
|
4MGE
 
 | | 1.85 Angstrom Resolution Crystal Structure of PTS System Cellobiose-specific Transporter Subunit IIB from Bacillus anthracis. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PTS system, ... | | Authors: | Minasov, G, Shuvalova, L, Halavaty, A, Winsor, J, Dubrovska, I, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-28 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Resolution Crystal Structure of PTS System Cellobiose-specific Transporter Subunit IIB from Bacillus anthracis.
TO BE PUBLISHED
|
|
4CKU
 
 | | Three dimensional structure of plasmepsin II in complex with hydroxyethylamine-based inhibitor | | Descriptor: | 5-[1,1-bis(oxidanylidene)-1,2-thiazinan-2-yl]-N3-[(2S,3R)-4-[2-(3-methoxyphenyl)propan-2-ylamino]-3-oxidanyl-1-phenyl-butan-2-yl]-N1,N1-dipropyl-benzene-1,3-dicarboxamide, PLASMEPSIN-2 | | Authors: | Tars, K, Leitans, J, Jaudzems, K. | | Deposit date: | 2014-01-08 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Plasmepsin Inhibitory Activity and Structure-Guided Optimization of a Potent Hydroxyethylamine-Based Antimalarial Hit.
Acs Med.Chem.Lett., 5, 2014
|
|
4BO4
 
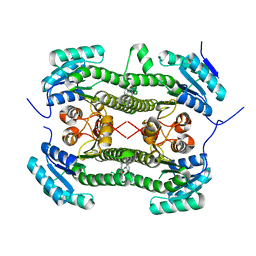 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with N-(2-methoxyphenyl)-3,4- dihydro-2H-quinoline-1-carboxamide at 2.7A resolution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, N-(2-methoxyphenyl)-3,4-dihydro-2H-quinoline-1-carboxamide | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-18 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4BP1
 
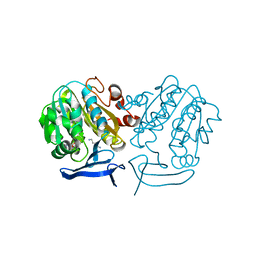 | | Crystal Structure of Plasmodium Falciparum Spermidine Synthase in Complex with 5'-Methylthioadenosine and Putrescine | | Descriptor: | 1,4-DIAMINOBUTANE, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, ... | | Authors: | Sprenger, J, Halander, J.C, Svensson, B, Al-Karadaghi, S, Persson, L. | | Deposit date: | 2013-05-23 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Three-Dimensional Structures of Plasmodium Falciparum Spermidine Synthase with Bound Inhibitors Suggest New Strategies for Drug Design
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4BOD
 
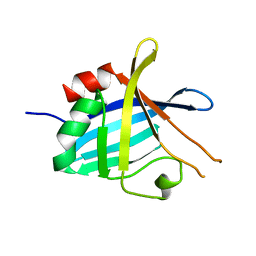 | | Complement regulator acquiring outer surface protein BbCRASP-4 or ErpC from Borrelia burgdorferi | | Descriptor: | ERPC | | Authors: | Brangulis, K, Petrovskis, I, Kazaks, A, Baumanis, V, Tars, K. | | Deposit date: | 2013-05-18 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal Structures of the Erp Protein Family Members Erpp and Erpc from Borrelia Burgdorferi Reveal the Reason for Different Affinities for Complement Regulator Factor H.
Biochim.Biophys.Acta, 1854, 2015
|
|
5WJJ
 
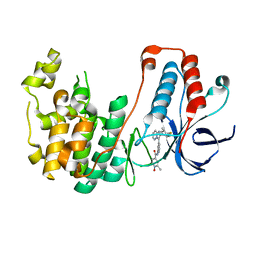 | | Structure-based Design, Synthesis, and Biological Evaluation of Imidazo[1,2-b]pyridazine-based p38 MAP Kinase Inhibitors | | Descriptor: | Mitogen-activated protein kinase 14, N-{4-[2-(4-fluoro-3-methylphenyl)imidazo[1,2-b]pyridazin-3-yl]pyridin-2-yl}-2-methyl-1-oxo-1lambda~5~-pyridine-4-carboxamide | | Authors: | Snell, G.P, Okada, K, Bragstad, K, Sang, B.-C. | | Deposit date: | 2017-07-23 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based design, synthesis, and biological evaluation of imidazo[1,2-b]pyridazine-based p38 MAP kinase inhibitors.
Bioorg. Med. Chem., 26, 2018
|
|
4MQB
 
 | | Crystal structure of thymidylate kinase from Staphylococcus aureus in complex with 2-(N-morpholino)ethanesulfonic acid | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, TETRAETHYLENE GLYCOL, Thymidylate kinase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2013-09-16 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of thymidylate kinase from Staphylococcus aureus in complex with 2-(N-morpholino)ethanesulfonic acid
To be Published
|
|
4MPB
 
 | | 1.7 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) from Staphylococcus aureus | | Descriptor: | Betaine aldehyde dehydrogenase, CHLORIDE ION, MAGNESIUM ION | | Authors: | Halavaty, A.S, Shuvalova, L, Minasov, G, Dubrovska, I, Winsor, J, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-12 | | Release date: | 2013-09-25 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based mutational studies of substrate inhibition of betaine aldehyde dehydrogenase BetB from Staphylococcus aureus.
Appl.Environ.Microbiol., 80, 2014
|
|
4MPY
 
 | | 1.85 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) from Staphylococcus aureus (IDP00699) in complex with NAD+ | | Descriptor: | Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Winsor, J, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-14 | | Release date: | 2013-10-09 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-based mutational studies of substrate inhibition of betaine aldehyde dehydrogenase BetB from Staphylococcus aureus.
Appl.Environ.Microbiol., 80, 2014
|
|
4CNT
 
 | | CRYSTAL STRUCTURE OF WT HUMAN CRMP-4 | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROPYRIMIDINASE-LIKE 3, SODIUM ION | | Authors: | Ponnusamy, R, Lohkamp, B. | | Deposit date: | 2014-01-24 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of Human Crmp-4: Correction of Intensities for Lattice-Translocation Disorder
Acta Crystallogr.,Sect.D, 70, 2014
|
|
