7A9A
 
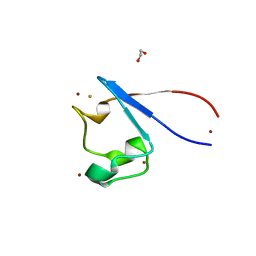 | | Crystal structure of rubredoxin B (Rv3250c) from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vakhrameev, D, Kavaleuski, A, Bukhdruker, S, Marin, E, Sushko, T, Grabovec, I.P, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2020-09-01 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | A new twist of rubredoxin function in M. tuberculosis.
Bioorg.Chem., 109, 2021
|
|
8G0E
 
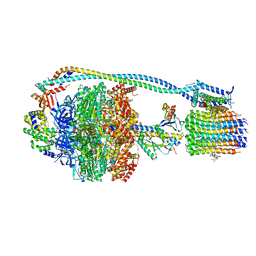 | | Cryo-EM structure of TBAJ-876-bound Mycobacterium smegmatis ATP synthase rotational state 3 | | Descriptor: | (1R,2S)-1-(6-bromo-2-methoxyquinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2,3,6-trimethoxypyridin-4-yl)butan-2-ol, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2023-01-31 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Mechanism of mycobacterial ATP synthase inhibition by squaramides and second generation diarylquinolines.
Embo J., 42, 2023
|
|
8G0D
 
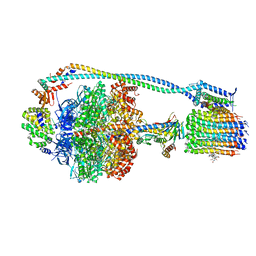 | | Cryo-EM structure of TBAJ-876-bound Mycobacterium smegmatis ATP synthase rotational state 2 (backbone model) | | Descriptor: | (1R,2S)-1-(6-bromo-2-methoxyquinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2,3,6-trimethoxypyridin-4-yl)butan-2-ol, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2023-01-31 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of mycobacterial ATP synthase inhibition by squaramides and second generation diarylquinolines.
Embo J., 42, 2023
|
|
6XNN
 
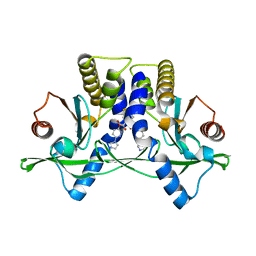 | | Crystal Structure of Mouse STING CTD complex with SR-717. | | Descriptor: | 4,5-difluoro-2-{[6-(1H-imidazol-1-yl)pyridazine-3-carbonyl]amino}benzoic acid, Stimulator of interferon genes protein | | Authors: | Chin, E.N, Yu, C, Wolan, D.W, Petrassi, H.M, Lairson, L.L. | | Deposit date: | 2020-07-03 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Antitumor activity of a systemic STING-activating non-nucleotide cGAMP mimetic.
Science, 369, 2020
|
|
4QDI
 
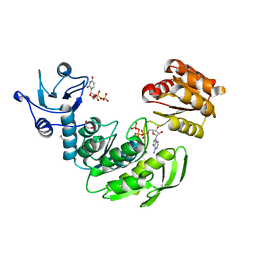 | | Crystal structure II of MurF from Acinetobacter baumannii | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | An, Y.J, Jeong, C.S, Cha, S.S. | | Deposit date: | 2014-05-13 | | Release date: | 2015-04-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ATP-binding mode including a carbamoylated lysine and two Mg(2+) ions, and substrate-binding mode in Acinetobacter baumannii MurF
Biochem.Biophys.Res.Commun., 450, 2014
|
|
8Z4R
 
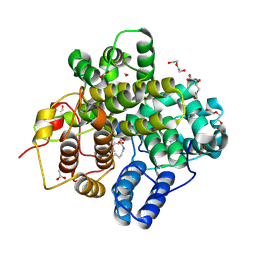 | | The crystal structure of a Hydroquinone Dioxygenase PaD with substrate | | Descriptor: | 2-methoxy-6-methyl-benzene-1,4-diol, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Liu, Z.W, Huang, J.-W, Wang, Y.X, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2024-04-17 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Substrate specificity of a branch of aromatic dioxygenases determined by three distinct motifs.
Nat Commun, 15, 2024
|
|
8IFP
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 1 | | Descriptor: | (1R,2S,5S)-3-[(2S)-2-(tert-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-6,6-dimethyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
7A50
 
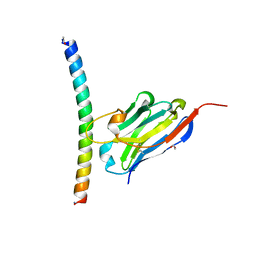 | | Crystal structure of the APH coiled-coil in complex with nanobody Nb26 | | Descriptor: | 1,2-ETHANEDIOL, Coiled-coil APH, Nanobody Nb26 | | Authors: | Hadzi, S. | | Deposit date: | 2020-08-20 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | A nanobody toolbox targeting dimeric coiled-coil modules for functionalization of designed protein origami structures.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8HXN
 
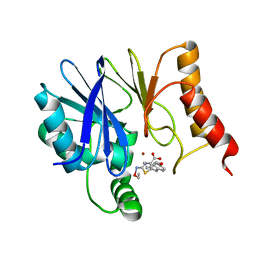 | | Crystal structure of B2 Sfh-I MBL in complex with 2-amino-5-(4-(but-3-en-1-yloxy)benzyl)thiazole-4-carboxylic acid | | Descriptor: | 2-azanyl-5-[(4-but-3-enoxyphenyl)methyl]-1,3-thiazole-4-carboxylic acid, Beta-lactamase, ETHANOL, ... | | Authors: | Yan, Y.-H, Zhu, K.-R, Li, G.-B. | | Deposit date: | 2023-01-05 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Discovery of 2-Aminothiazole-4-carboxylic Acids as Broad-Spectrum Metallo-beta-lactamase Inhibitors by Mimicking Carbapenem Hydrolysate Binding.
J.Med.Chem., 66, 2023
|
|
6MR5
 
 | |
6ZOE
 
 | | AcrB-F563A symmetric T protomer | | Descriptor: | 1,2-ETHANEDIOL, DARPIN, DECANE, ... | | Authors: | Tam, H.K, Foong, W.E, Pos, K.M. | | Deposit date: | 2020-07-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Allosteric drug transport mechanism of multidrug transporter AcrB.
Nat Commun, 12, 2021
|
|
9FKB
 
 | | Tail of emppty Haloferax tailed virus 1 | | Descriptor: | Baseplate to tube adapter protein gp41, HK97 gp6-like/SPP1 gp15-like head-tail connector, MAGNESIUM ION, ... | | Authors: | Zhang, D, Daum, B, Isupov, M.N, McLaren, M. | | Deposit date: | 2024-06-03 | | Release date: | 2025-06-18 | | Last modified: | 2025-10-22 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Cryo-EM resolves the structure of the archaeal dsDNA virus HFTV1 from head to tail.
Sci Adv, 11, 2025
|
|
8WAG
 
 | | Crystal structure of the C-terminal fragment (residues 716-982) of Arabidopsis thaliana CHUP1 | | Descriptor: | Protein CHUP1, chloroplastic | | Authors: | Shimada, A, Nakamura, Y, Takano, A, Kohda, D. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | CHLOROPLAST UNUSUAL POSITIONING 1 is a plant-specific actin polymerization factor regulating chloroplast movement.
Plant Cell, 36, 2024
|
|
6N2G
 
 | | Crystal structure of Caenorhabditis elegans NAP1 | | Descriptor: | Nucleosome Assembly Protein | | Authors: | Bhattacharyya, S, DArcy, S. | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Characterization of Caenorhabditis elegans Nucleosome Assembly Protein 1 Uncovers the Role of Acidic Tails in Histone Binding.
Biochemistry, 58, 2019
|
|
7P51
 
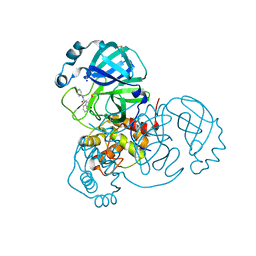 | | CRYSTAL STRUCTURE OF THE SARS-COV-2 MAIN PROTEASE COMPLEXED WITH FRAGMENT F01 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(5-chloropyridin-2-yl)-3-oxo-2,3-dihydro-1H-indene-1-carboxamide, ... | | Authors: | Hanoulle, X, Moschidi, D. | | Deposit date: | 2021-07-13 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.474 Å) | | Cite: | NMR Spectroscopy of the Main Protease of SARS-CoV-2 and Fragment-Based Screening Identify Three Protein Hotspots and an Antiviral Fragment.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7OMB
 
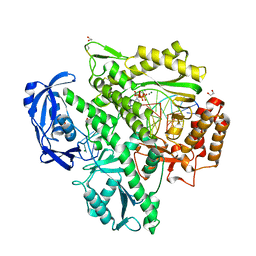 | | Crystal structure of KOD DNA Polymerase in a ternary complex with a p/t duplex containing an extended 5' single stranded template overhang | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA polymerase, ... | | Authors: | Betz, K, Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2021-05-21 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Basis for The Recognition of Deaminated Nucleobases by An Archaeal DNA Polymerase.
Chembiochem, 22, 2021
|
|
6ZY7
 
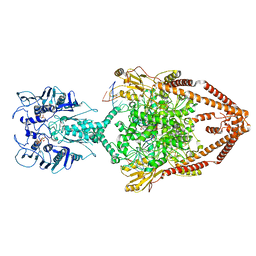 | | Cryo-EM structure of the entire Human topoisomerase II alpha in State 1 | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(*CP*GP*CP*GP*CP*AP*TP*CP*GP*TP*CP*AP*TP*CP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*GP*AP*TP*GP*AP*CP*GP*AP*TP*G)-3'), ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2020-07-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.64 Å) | | Cite: | Structural basis for allosteric regulation of Human Topoisomerase II alpha.
Nat Commun, 12, 2021
|
|
7R2G
 
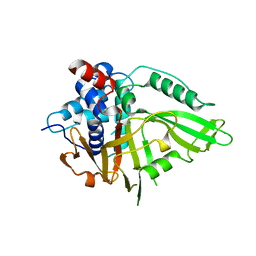 | | USP15 D1D2 in catalytically-competent state bound to mitoxantrone stack (isoform 2) | | Descriptor: | 1,4-DIHYDROXY-5,8-BIS({2-[(2-HYDROXYETHYL)AMINO]ETHYL}AMINO)-9,10-ANTHRACENEDIONE, GLYCEROL, Ubiquitin carboxyl-terminal hydrolase 15, ... | | Authors: | Priyanka, A, Sixma, T.K. | | Deposit date: | 2022-02-04 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Mitoxantrone stacking does not define the active or inactive state of USP15 catalytic domain.
J.Struct.Biol., 214, 2022
|
|
6MVL
 
 | |
5LNC
 
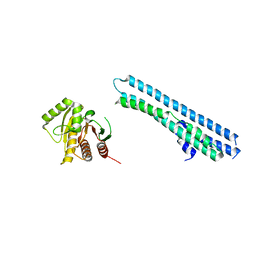 | |
9JFL
 
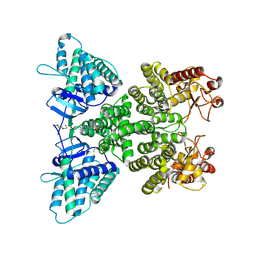 | |
7OQP
 
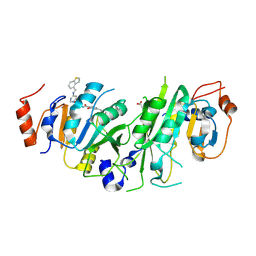 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ113 | | Descriptor: | ACETATE ION, N-[[(3R)-1-[6-(1-benzothiophen-4-ylmethylamino)pyrimidin-4-yl]-3-oxidanyl-piperidin-3-yl]methyl]-2-oxidanyl-4-[[(3S)-3-propan-2-yl-2-azaspiro[3.3]heptan-2-yl]methyl]benzamide, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-06-03 | | Release date: | 2021-10-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
8TJM
 
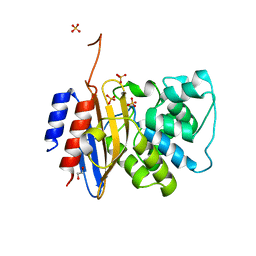 | | Crystal structure of KPC-44 carbapenemase | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, beta-lactamase | | Authors: | Sun, Z, Palzkill, T, Hu, L, Lin, H, Sankaran, B, Wang, J, Prasad, B. | | Deposit date: | 2023-07-23 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Klebsiella pneumoniae carbapenemase variant 44 acquires ceftazidime-avibactam resistance by altering the conformation of active-site loops.
J.Biol.Chem., 300, 2023
|
|
8R85
 
 | | Xylanase from Bacillus circulans mutant E78Q/W9A | | Descriptor: | DI(HYDROXYETHYL)ETHER, Endo-1,4-beta-xylanase, GLYCEROL | | Authors: | Chikunova, A, Saberi, M, Ubbink, M. | | Deposit date: | 2023-11-28 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Bimodal substrate binding in the active site of the glycosidase BcX.
Febs J., 291, 2024
|
|
8ZB5
 
 | | Crystal structure of NudC from Mycobacterium abscessus in complex with AMP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE MONOPHOSPHATE, CALCIUM ION, ... | | Authors: | Meng, L, Zhang, Y, Xu, J, Liu, J. | | Deposit date: | 2024-04-26 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Studies on Mycobacterial NudC Reveal a Class of Zinc Independent NADH Pyrophosphatase.
J.Mol.Biol., 436, 2024
|
|
