8QEL
 
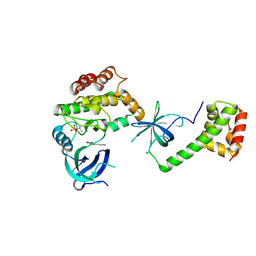 | | PKR kinase domain- eIF2alpha in complex with compound | | Descriptor: | (3~{Z})-3-[(4-methyl-1~{H}-imidazol-5-yl)methylidene]-2-oxidanylidene-1~{H}-indole-5-carboxamide, Eukaryotic translation initiation factor 2 subunit alpha, Interferon-induced, ... | | Authors: | Nawrotek, A, Vuillard, L, Miallau, L. | | Deposit date: | 2023-08-31 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | PKR kinase domain- eIF2alpha in complex with compound
To Be Published
|
|
6R1L
 
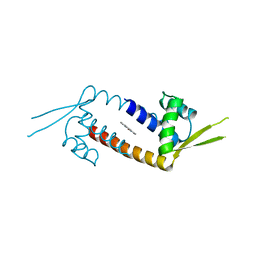 | |
8G20
 
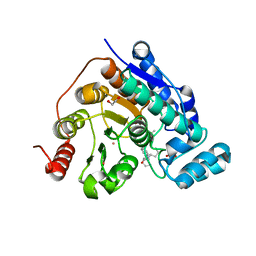 | |
6MY5
 
 | |
9AVJ
 
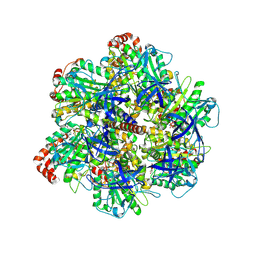 | | PS3 F1 ATPase Wild type | | Descriptor: | ATP synthase gamma chain, ATP synthase subunit alpha, ATP synthase subunit beta, ... | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2024-03-03 | | Release date: | 2024-10-30 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | The molecular structure of an axle-less F 1 -ATPase.
Biochim Biophys Acta Bioenerg, 1866, 2024
|
|
7Q80
 
 | |
6WP4
 
 | | Pyruvate Kinase M2 mutant-S37E | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Nandi, S, Razzaghi, M, Srivastava, D, Dey, M. | | Deposit date: | 2020-04-26 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for allosteric regulation of pyruvate kinase M2 by phosphorylation and acetylation.
J.Biol.Chem., 295, 2020
|
|
6ON3
 
 | | A substrate bound structure of L-DOPA dioxygenase from Streptomyces sclerotialus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,4-DIHYDROXYPHENYLALANINE, FE (II) ION, ... | | Authors: | Wang, Y, Shin, I, Fu, Y, Colabroy, K, Liu, A. | | Deposit date: | 2019-04-19 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structures of L-DOPA Dioxygenase fromStreptomyces sclerotialus.
Biochemistry, 58, 2019
|
|
6UG2
 
 | | C2 symmetric peptide design number 1, Zappy, crystal form 2 | | Descriptor: | C2-1, Zappy, crystal form 2, ... | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-25 | | Release date: | 2020-12-02 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6X1Y
 
 | | Mre11 dimer in complex with small molecule modulator PFMI | | Descriptor: | (5Z)-5-[(3-methoxyphenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclease SbcCD subunit D | | Authors: | Arvai, A.S, Moiani, D, Tainer, J.A. | | Deposit date: | 2020-05-19 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Fragment- and structure-based drug discovery for developing therapeutic agents targeting the DNA Damage Response.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
6OOY
 
 | | Asymmetric hTNF-alpha | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Tumor necrosis factor, {1-[(2,5-dimethylphenyl)methyl]-1H-benzimidazol-2-yl}methanol | | Authors: | Arakaki, T.L, Edwards, T.E, Ceska, T. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Small molecules that inhibit TNF signalling by stabilising an asymmetric form of the trimer.
Nat Commun, 10, 2019
|
|
7BBO
 
 | | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group P212121 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-18 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group P21212
To Be Published
|
|
7RFP
 
 | | Mouse GITR (mGITR) with DTA-1 Fab fragment | | Descriptor: | DTA-1 (heavy chain), DTA-1 (light chain), Tumor necrosis factor receptor superfamily member 18,Enhanced green fluorescent protein | | Authors: | Meyerson, J.R, He, C. | | Deposit date: | 2021-07-14 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Therapeutic antibody activation of the glucocorticoid-induced TNF receptor by a clustering mechanism.
Sci Adv, 8, 2022
|
|
8AXS
 
 | | Sialidases and Fucosidases of Akkermansia muciniphila are key for rapid growth on colonic mucin and nutrient sharing amongst mucin-associated human gut microbiota | | Descriptor: | 1,2-ETHANEDIOL, 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CALCIUM ION, ... | | Authors: | Sakanaka, H, Nielsen, T.S, Pichler, M.J, Nordberg Karlsson, E, Abou Hachem, M, Morth, J.P. | | Deposit date: | 2022-08-31 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Sialidases and fucosidases of Akkermansia muciniphila are crucial for growth on mucin and nutrient sharing with mucus-associated gut bacteria.
Nat Commun, 14, 2023
|
|
8J7Z
 
 | | Structure of FCP trimer in Cyclotella meneghiniana | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, CHLOROPHYLL A, ... | | Authors: | Shen, L.L, Li, Z.H, Shen, J.R, Wang, W.D. | | Deposit date: | 2023-04-28 | | Release date: | 2023-12-20 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structural insights into photosystem II supercomplex and trimeric FCP antennae of a centric diatom Cyclotella meneghiniana.
Nat Commun, 14, 2023
|
|
6XFS
 
 | | Class C beta-lactamase from Escherichia coli in complex with Tazobactam | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-16 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Class C beta-lactamase from Escherichia coli in complex with Tazobactam
To Be Published
|
|
7RP2
 
 | | Crystal structure of Kas G12C in complex with 2H11 CLAMP | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, GTPase KRas, ... | | Authors: | Oh, A, Tam, C, Wang, W. | | Deposit date: | 2021-08-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformation-locking antibodies for the discovery and characterization of KRAS inhibitors.
Nat.Biotechnol., 40, 2022
|
|
8BAQ
 
 | | E. coli C7 DarT1 in complex with NAD+ | | Descriptor: | 1,2-ETHANEDIOL, DarT ssDNA thymidine ADP-ribosyltransferase family protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2022-10-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the reversible ADP-ribosylation of guanosine bases.
Mol.Cell, 83, 2023
|
|
8BAU
 
 | | Phytophthora nicotianae var. parasitica NADAR in complex with ADP-ribose | | Descriptor: | 1,2-ETHANEDIOL, NADAR domain-containing protein, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2022-10-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis for the reversible ADP-ribosylation of guanosine bases.
Mol.Cell, 83, 2023
|
|
6WOI
 
 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with 1-Diphosphoinositol pentakisphosphate, Mg, and Fluoride ion, presoaked with 1,5-IP8, Mg and Fluoride for 30 seconds | | Descriptor: | (1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Zong, G.N, Wang, H.C, Shears, S.B. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | New structural insights reveal an expanded reaction cycle for inositol pyrophosphate hydrolysis by human DIPP1.
Faseb J., 35, 2021
|
|
7QUB
 
 | | EV-A71-3Cpro in complex with inhibitor MG78 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, Protease 3C, SODIUM ION | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-01-17 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | From Repurposing to Redesign: Optimization of Boceprevir to Highly Potent Inhibitors of the SARS-CoV-2 Main Protease.
Molecules, 27, 2022
|
|
7QGZ
 
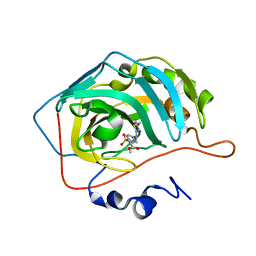 | | Human carbonic anhydrase II in complex with Methyl 3-((4-methylthiazol-2-yl)(4-sulfamoylphenyl)amino)propanoate | | Descriptor: | Carbonic anhydrase 2, ZINC ION, methyl 3-[(4-methyl-1,3-thiazol-2-yl)-(4-sulfamoylphenyl)amino]propanoate | | Authors: | Paketuryte-Latve, V, Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-12-10 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Beta and Gamma Amino Acid-Substituted Benzenesulfonamides as Inhibitors of Human Carbonic Anhydrases.
Pharmaceuticals, 15, 2022
|
|
6OGN
 
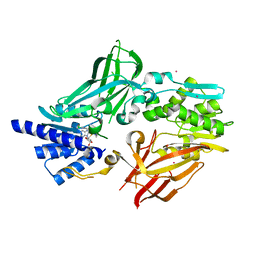 | | Crystal structure of mouse protein arginine methyltransferase 7 in complex with SGC8158 chemical probe | | Descriptor: | 5'-S-(4-{[(4'-chloro[1,1'-biphenyl]-3-yl)methyl]amino}butyl)-5'-thioadenosine, Protein arginine N-methyltransferase 7, UNKNOWN ATOM OR ION, ... | | Authors: | Halabelian, L, Dong, A, Zeng, H, Li, Y, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-03 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pharmacological inhibition of PRMT7 links arginine monomethylation to the cellular stress response.
Nat Commun, 11, 2020
|
|
7QL8
 
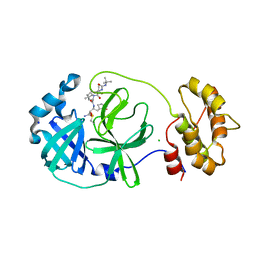 | | SARS-COV2 Main Protease in complex with inhibitor MG78 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2021-12-19 | | Release date: | 2022-04-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.807 Å) | | Cite: | From Repurposing to Redesign: Optimization of Boceprevir to Highly Potent Inhibitors of the SARS-CoV-2 Main Protease.
Molecules, 27, 2022
|
|
7T1K
 
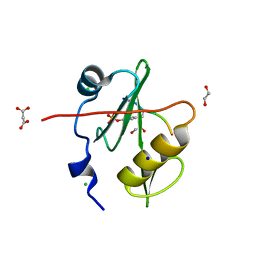 | | Crystal structure of a superbinder Fes SH2 domain (sFes1) in complex with a high affinity phosphopeptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MALONATE ION, ... | | Authors: | Martyn, G.D, Singer, A.U, Veggiani, G, Kurinov, I, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2021-12-02 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Engineered SH2 Domains for Targeted Phosphoproteomics.
Acs Chem.Biol., 17, 2022
|
|
