6HQ0
 
 | | Crystal structure of ENL (MLLT1), apo form | | Descriptor: | 1,2-ETHANEDIOL, Protein ENL | | Authors: | Heidenreich, D, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure-Based Approach toward Identification of Inhibitory Fragments for Eleven-Nineteen-Leukemia Protein (ENL).
J.Med.Chem., 61, 2018
|
|
7SSE
 
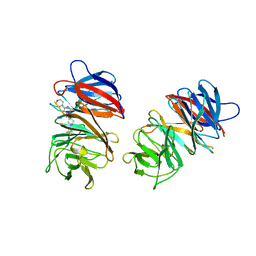 | | Crystal structure of the WDR domain of human DCAF1 in complex with CYCA-117-70 | | Descriptor: | DDB1- and CUL4-associated factor 1, N-[(3R)-1-(3-fluorophenyl)piperidin-3-yl]-6-(morpholin-4-yl)pyrimidin-4-amine | | Authors: | Kimani, S, Owen, J, Li, A, Dong, A, Li, Y, Hutchinson, A, Seitova, A, Shahani, V.M, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-11-10 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery of a Novel DCAF1 Ligand Using a Drug-Target Interaction Prediction Model: Generalizing Machine Learning to New Drug Targets.
J.Chem.Inf.Model., 63, 2023
|
|
8UVC
 
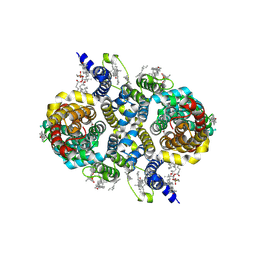 | | Structure of NaDC3-aKG complex | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-OXOGLUTARIC ACID, CHOLESTEROL, ... | | Authors: | Li, Y, Wang, D.N, Mindell, J.A, Rice, W.J, Song, J, Mikusevic, V, Marden, J.J, Becerril, A, Kuang, H, Wang, B. | | Deposit date: | 2023-11-02 | | Release date: | 2024-12-25 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (2.09 Å) | | Cite: | Substrate translocation and inhibition in human dicarboxylate transporter NaDC3.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8UVE
 
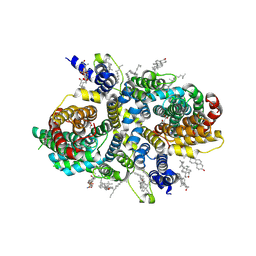 | | Structure of NaDC3-Succinate complex in Coo-Ci conformation | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL, Na(+)/dicarboxylate cotransporter 3, ... | | Authors: | Li, Y, Wang, D.N, Mindell, J.A, Rice, W.J, Song, J, Mikusevic, V, Marden, J.J, Becerril, A, Kuang, H, Wang, B. | | Deposit date: | 2023-11-02 | | Release date: | 2024-12-25 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Substrate translocation and inhibition in human dicarboxylate transporter NaDC3.
Nat.Struct.Mol.Biol., 32, 2025
|
|
7PRC
 
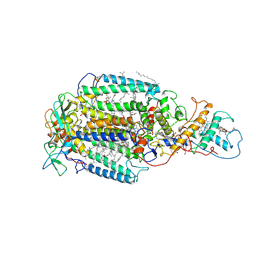 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOPSEUDOMONAS VIRIDIS (DG-420315 (TRIAZINE) COMPLEX) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, 2-CHLORO-4-ETHYLAMINO-6-(R(+)-2'-CYANO-4-BUTYLAMINO)-1,3,5-TRIAZINE, BACTERIOCHLOROPHYLL B, ... | | Authors: | Lancaster, C.R.D, Michel, H. | | Deposit date: | 1997-08-01 | | Release date: | 1999-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Refined crystal structures of reaction centres from Rhodopseudomonas viridis in complexes with the herbicide atrazine and two chiral atrazine derivatives also lead to a new model of the bound carotenoid.
J.Mol.Biol., 286, 1999
|
|
8UVF
 
 | | Structure of NaDC3-DMS complex in Ci-Ci conformation | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL, Na(+)/dicarboxylate cotransporter 3, ... | | Authors: | Li, Y, Wang, D.N, Mindell, J.A, Rice, W.J, Song, J, Mikusevic, V, Marden, J.J, Becerril, A, Kuang, H, Wang, B. | | Deposit date: | 2023-11-02 | | Release date: | 2024-12-25 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | Substrate translocation and inhibition in human dicarboxylate transporter NaDC3.
Nat.Struct.Mol.Biol., 32, 2025
|
|
6NQT
 
 | | GalNac-T2 soaked with UDP-sugar | | Descriptor: | MANGANESE (II) ION, Polypeptide N-acetylgalactosaminyltransferase 2, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{R},4~{R},5~{R},6~{R})-3-(hex-5-ynoylamino)-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-yl] hydrogen phosphate | | Authors: | Fernandez, D, Bertozzi, C.R, Schumann, B, Agbay, A. | | Deposit date: | 2019-01-21 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Bump-and-Hole Engineering Identifies Specific Substrates of Glycosyltransferases in Living Cells.
Mol.Cell, 78, 2020
|
|
6WYR
 
 | | Crystal structure of anti-Muscle Specific Kinase (MuSK) Fab, MuSK1A | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MuSK1A heavy chain, ... | | Authors: | Vieni, C, Ekiert, D. | | Deposit date: | 2020-05-13 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Affinity maturation is required for pathogenic monovalent IgG4 autoantibody development in myasthenia gravis.
J.Exp.Med., 217, 2020
|
|
6QNK
 
 | | Antibody FAB fragment targeting Gi protein heterotrimer | | Descriptor: | 1,2-ETHANEDIOL, D-MALATE, FAB heavy chain, ... | | Authors: | Tsai, C.-J, Muehle, J, Pamula, F, Dawson, R.J.P, Maeda, S, Deupi, X, Schertler, G.F.X. | | Deposit date: | 2019-02-11 | | Release date: | 2019-07-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cryo-EM structure of the rhodopsin-G alpha i-beta gamma complex reveals binding of the rhodopsin C-terminal tail to the G beta subunit.
Elife, 8, 2019
|
|
8GAI
 
 | |
8JK2
 
 | | Crystal structure of QF-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-31 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.742 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
6NFL
 
 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B with loop 7 from APOBEC3G complexed with 2-HP | | Descriptor: | 1,2-ETHANEDIOL, 1,3-diazinan-2-one, CHLORIDE ION, ... | | Authors: | Shi, K, Orellana, K, Aihara, H. | | Deposit date: | 2018-12-20 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.731 Å) | | Cite: | Active site plasticity and possible modes of chemical inhibition of the human DNA deaminase APOBEC3B
Faseb Bioadv, 2, 2020
|
|
6NUN
 
 | |
8PJI
 
 | | MLLT1 in complex with compound 10a | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Protein ENL, ... | | Authors: | Raux, B, Diaz-Saez, L, Huber, K.V.M, Fedorov, O, Owen, D.R, Londregan, A.T, Bountra, C, Edwards, A, Arrowsmith, C. | | Deposit date: | 2023-06-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of PFI-6, a small-molecule chemical probe for the YEATS domain of MLLT1 and MLLT3.
Bioorg.Med.Chem.Lett., 98, 2023
|
|
7BOM
 
 | | Crystal structure of recombinant horse spleen apo-R52C/E56C/R59C/E63C-Fr immobilized with gold ions. | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Hishikawa, Y, Maity, B, Ito, N, Abe, S, Lu, D, Ueno, T. | | Deposit date: | 2020-03-19 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Design of Multinuclear Gold Binding Site at the Two-fold Symmetric Interface of the Ferritin Cage
Chem Lett., 49, 2021
|
|
8FEY
 
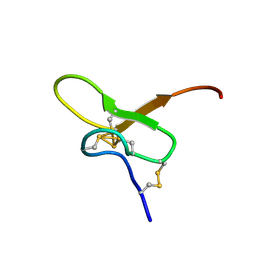 | | Solution structure of Pmu1a | | Descriptor: | Pmu1a toxin | | Authors: | Daly, N.L, Wilson, D.T. | | Deposit date: | 2022-12-07 | | Release date: | 2023-03-29 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Pmu1a, a novel spider toxin with dual inhibitory activity at pain targets hNa V 1.7 and hCa V 3 voltage-gated channels.
Febs J., 290, 2023
|
|
8UAV
 
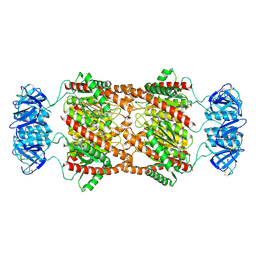 | | Cryo-EM Structure of Brucella Abortus Lumazine Synthase (BLS) Engineered with Shiga Toxin I subunit B (Stx1B) | | Descriptor: | Shiga toxin subunit B,6,7-dimethyl-8-ribityllumazine synthase 2 | | Authors: | Cristofalo, A.E, Sharma, A, Cerutti, M.L, Sharma, K, Zylberman, V, Goldbaum, F.A, Borgnia, M.J, Otero, L.H. | | Deposit date: | 2023-09-22 | | Release date: | 2025-03-26 | | Last modified: | 2025-10-08 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | Cryo-EM structures of engineered Shiga toxin-based immunogens capable of eliciting neutralizing antibodies with therapeutic potential against hemolytic uremic syndrome.
Protein Sci., 34, 2025
|
|
7SPP
 
 | |
6Z57
 
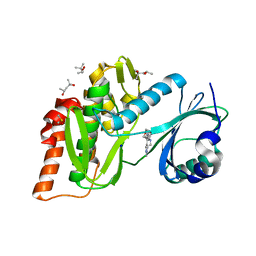 | | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2004078 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 10-(2-morpholin-4-ylethyl)-7-oxa-10,13,17,18,21-pentazatetracyclo[12.5.2.12,6.017,20]docosa-1(20),2(22),3,5,14(21),15,18-heptaene, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2004078
To Be Published
|
|
9B1B
 
 | | EV-D68 in complex with inhibitor Jun11-78-7 | | Descriptor: | 4-[(propan-2-yl)oxy]-N-{(4M)-4-[1-(2,2,2-trifluoroethyl)-1H-pyrazol-4-yl]quinolin-8-yl}benzamide, Capsid protein VP1, Capsid protein VP4, ... | | Authors: | Klose, T, Kuhn, R.J, Jun, W, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-03-13 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Design of enterovirus D68 capsid inhibitors with in vivo antiviral efficacy
To Be Published
|
|
9B18
 
 | |
6NOB
 
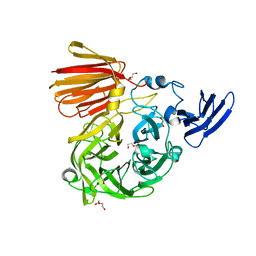 | |
8OWG
 
 | | Crystal structure of D1228V c-MET bound by compound 2 | | Descriptor: | 5-[3,5-bis(fluoranyl)phenyl]-1-[(1S)-1-phenylethyl]pyrimidine-2,4-dione, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-27 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.631 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
9B2Q
 
 | | Crystal Structure of Pantothenate Synthetase from Candida albicans. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Regan, J, Avad, K.A, Alaidi, O, Seetharaman, J, Palmer, G.E, Hevener, K.E. | | Deposit date: | 2024-03-15 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small molecule inhibitors of fungal pantothenate synthetase can provide a valid and potentially efficacious strategy for antifungal development.
To Be Published
|
|
6NR0
 
 | |
