8YGV
 
 | | F1 domain of Non-catalytic site depleted and epsilon C-terminal domain deleted FoF1-ATPase from Bacillus PS3,nucleotide depleted condition | | Descriptor: | ATP synthase gamma chain, ATP synthase subunit alpha, ATP synthase subunit beta | | Authors: | Kobayashi, R, Nakano, A, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2024-02-27 | | Release date: | 2025-03-05 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | ADP-inhibited structure of non-catalytic site-depleted F o F 1 -ATPase from thermophilic Bacillus sp. PS-3.
Biochim Biophys Acta Bioenerg, 1866, 2025
|
|
6PTM
 
 | |
6BIA
 
 | | Crystal structure of Ps i-CgsB | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Hettle, A.G, Boraston, A.B. | | Deposit date: | 2017-11-01 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Molecular Basis of Polysaccharide Sulfatase Activity and a Nomenclature for Catalytic Subsites in this Class of Enzyme.
Structure, 26, 2018
|
|
8YH8
 
 | | F1 domain of Non-catalytic site depleted and epsilon C-terminal domain deleted FoF1-ATPase from Bacillus PS3,under ATP saturated condition | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Kobayashi, R, Nakano, A, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2024-02-27 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | ADP-inhibited structure of non-catalytic site-depleted F o F 1 -ATPase from thermophilic Bacillus sp. PS-3.
Biochim Biophys Acta Bioenerg, 1866, 2025
|
|
1R3G
 
 | | 1.16A X-ray structure of the synthetic DNA fragment with the incorporated 2'-O-[(2-Guanidinium)ethyl]-5-methyluridine residues | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(GMU)P*AP*CP*GP*C)-3'), MAGNESIUM ION | | Authors: | Prakash, T.P, Puschl, A, Lesnik, E, Tereshko, V, Egli, M, Manoharan, M. | | Deposit date: | 2003-10-01 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | 2'-O-[2-(Guanidinium)ethyl]-Modified Oligonucleotides: Stabilizing Effect on Duplex and Triplex Structures.
Org.Lett., 6, 2004
|
|
7FJ0
 
 | |
7FHI
 
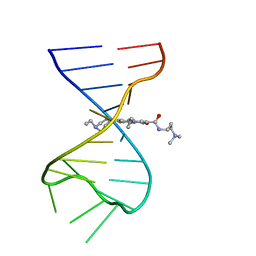 | |
1JZV
 
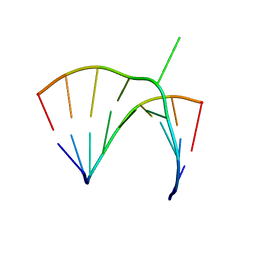 | | Crystal structure of a bulged RNA from the SL2 stem-loop of the HIV-1 psi-RNA | | Descriptor: | 5'-R(*CP*AP*GP*UP*AP*CP*GP*(5IC)P*C)-3', 5'-R(*GP*GP*CP*GP*AP*CP*(5BU)P*G)-3', MAGNESIUM ION | | Authors: | Xiong, Y, Sudarsanakumar, C, Deng, J, Pan, B, Sundaralingam, M. | | Deposit date: | 2001-09-17 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a Bulged RNA from the SL2 Stem-loop of the HIV-1 psi-RNA
To be Published
|
|
6BLJ
 
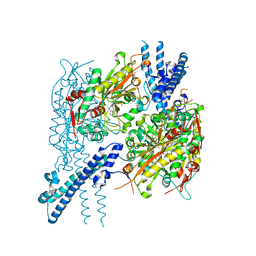 | |
1O3Z
 
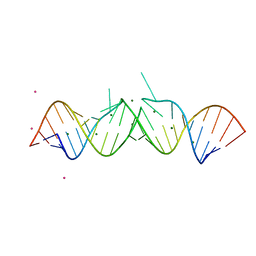 | | HIV-1 DIS(MAL) DUPLEX RU HEXAMINE-SOAKED | | Descriptor: | HIV-1 DIS(MAL) GENOMIC RNA, MAGNESIUM ION, RUTHENIUM ION | | Authors: | Ennifar, E, Walter, P, Dumas, P. | | Deposit date: | 2003-05-16 | | Release date: | 2003-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A crystallographic study of the binding of 13 metal ions to two related RNA duplexes.
Nucleic Acids Res., 31, 2003
|
|
1MSY
 
 | |
1K29
 
 | | Solution Structure of a DNA Duplex Containing M1G Opposite a 2 Base Pair Deletion | | Descriptor: | 5'-D(*AP*TP*CP*GP*CP*(M1G)P*CP*GP*GP*CP*AP*TP*G)-3', 5'-D(*CP*AP*TP*GP*CP*CP*GP*CP*GP*AP*T)-3' | | Authors: | Schnetz-Boutaud, N.C, Saleh, S, Marnett, L.J, Stone, M.P. | | Deposit date: | 2001-09-26 | | Release date: | 2002-01-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The exocyclic 1,N2-deoxyguanosine pyrimidopurinone M1G is a chemically stable DNA adduct when placed opposite a two-base deletion in the (CpG)3 frameshift hotspot of the Salmonella typhimurium hisD3052 gene.
Biochemistry, 40, 2001
|
|
1FIX
 
 | |
6G38
 
 | | Crystal structure of haspin in complex with tubercidin | | Descriptor: | '2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Heroven, C, Chaikuad, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-24 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Halogen-Aromatic pi Interactions Modulate Inhibitor Residence Times.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6G3C
 
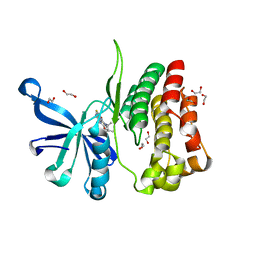 | | Crystal Structure of JAK2-V617F pseudokinase domain in complex with Compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[3,5-bis(fluoranyl)-4-oxidanyl-phenyl]amino]-5,7,7-trimethyl-8-(3-methylbutyl)pteridin-6-one, Tyrosine-protein kinase | | Authors: | Dekker, C, Hinniger, A. | | Deposit date: | 2018-03-24 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
Acs Chem.Biol., 14, 2019
|
|
6UXX
 
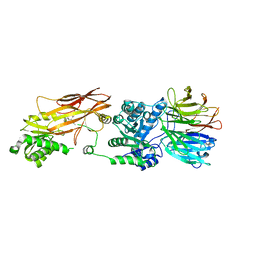 | | PRMT5:MEP50 Complexed with Allosteric Inhibitor Compound 1a | | Descriptor: | (5R)-2-amino-5-(4-methoxyphenyl)-3-methyl-5-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-yl]-3,5-dihydro-4H-imidazol-4-one, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | Palte, R.L, Schneider, S.E. | | Deposit date: | 2019-11-08 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Allosteric Modulation of Protein Arginine Methyltransferase 5 (PRMT5).
Acs Med.Chem.Lett., 11, 2020
|
|
8Z5J
 
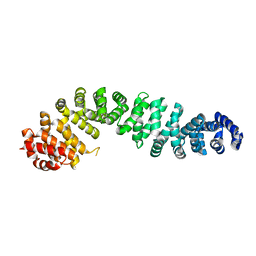 | | Beta-catenin Crystal Structure | | Descriptor: | Catenin beta-1 | | Authors: | Tim, F. | | Deposit date: | 2024-04-18 | | Release date: | 2024-05-22 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Novel 1-Phenylpiperidine Urea-Containing Derivatives Inhibiting beta-Catenin/BCL9 Interaction and Exerting Antitumor Efficacy through the Activation of Antigen Presentation of cDC1 Cells.
J.Med.Chem., 67, 2024
|
|
6V7I
 
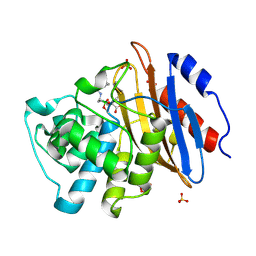 | | Structure of KPC-2 bound to Vaborbactam at 1.25 A | | Descriptor: | Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, SULFATE ION, ... | | Authors: | Pemberton, O.A, Chen, Y. | | Deposit date: | 2019-12-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis and Binding Kinetics of Vaborbactam in Class A beta-Lactamase Inhibition.
Antimicrob.Agents Chemother., 64, 2020
|
|
8YRF
 
 | |
6UZR
 
 | |
8QJ1
 
 | |
6ZT9
 
 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
6ZT7
 
 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
6WTO
 
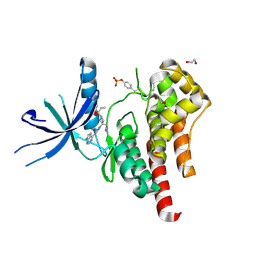 | | Human JAK2 JH1 domain in complex with Baricitinib | | Descriptor: | 1,2-ETHANEDIOL, Baricitinib, Tyrosine-protein kinase JAK2 | | Authors: | Yu, S, Nithianantham, S, Fischer, M. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Degradation of Janus kinases in CRLF2-rearranged acute lymphoblastic leukemia.
Blood, 138, 2021
|
|
8JQR
 
 | | Structure of human TRPV1 in complex with antagonist | | Descriptor: | 4-(7-Hydroxy-2-isopropyl-4-oxoquinazolin-3(4H)-yl)benzonitrile, CHOLESTEROL, Transient receptor potential cation channel subfamily V member 1,PreScission Site,Green fluorescent protein | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2023-06-14 | | Release date: | 2024-08-14 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structural basis of TRPV1 inhibition by SAF312 and cholesterol.
Nat Commun, 15, 2024
|
|
