6EAM
 
 | |
6EDU
 
 | |
4G7O
 
 | |
4GAW
 
 | | Crystal structure of active human granzyme H | | Descriptor: | CHLORIDE ION, Granzyme H, SULFATE ION | | Authors: | Wang, L, Li, Q, Wu, L, Zhang, K, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2012-07-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification of SERPINB1 as a physiological inhibitor of human granzyme H
J.Immunol., 190, 2013
|
|
4OSD
 
 | | Dimer of a C-terminal fragment of phage T4 gp5 beta-helix | | Descriptor: | 9-OCTADECENOIC ACID, MAGNESIUM ION, PALMITIC ACID, ... | | Authors: | Buth, S.A, Leiman, P.G, Shneider, M.M. | | Deposit date: | 2014-02-12 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure and Biophysical Properties of a Triple-Stranded Beta-Helix Comprising the Central Spike of Bacteriophage T4.
Viruses, 7, 2015
|
|
4OX9
 
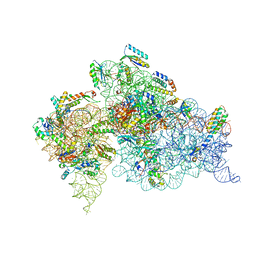 | | Crystal structure of the aminoglycoside resistance methyltransferase NpmA bound to the 30S ribosomal subunit | | Descriptor: | 16S rRNA, 16S rRNA (adenine(1408)-N(1))-methyltransferase, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Conn, G.L, Dunham, C.M. | | Deposit date: | 2014-02-04 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.8035 Å) | | Cite: | Molecular recognition and modification of the 30S ribosome by the aminoglycoside-resistance methyltransferase NpmA.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4FW7
 
 | |
4FZB
 
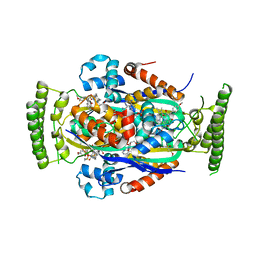 | | Structure of thymidylate synthase ThyX complexed to a new inhibitor | | Descriptor: | 2-hydroxy-3-(4-methoxybenzyl)naphthalene-1,4-dione, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Basta, T, Boum, Y, Briffotaux, J, Becker, H.F, Lamarre-Jouenne, I, Lambry, J.C, Skouloubris, S, Liebl, U, van Tilbeurgh, H, Graille, M, Myllylkallio, H. | | Deposit date: | 2012-07-06 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Mechanistic and structural basis for inhibition of thymidylate synthase ThyX.
Open Biology, 2, 2012
|
|
4OKN
 
 | | Crystal structure of human muscle L-lactate dehydrogenase, ternary complex with NADH and oxalate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, KANAMYCIN A, L-lactate dehydrogenase A chain, ... | | Authors: | Kolappan, S, Craig, L. | | Deposit date: | 2014-01-22 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of lactate dehydrogenase A (LDHA) in apo, ternary and inhibitor-bound forms.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4DV6
 
 | | Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, A915G | | Descriptor: | 16S rRNA, MAGNESIUM ION, ZINC ION, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-22 | | Release date: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (3.297 Å) | | Cite: | A structural basis for streptomycin resistance
To be Published
|
|
4DR6
 
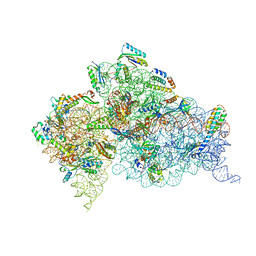 | | Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with codon, near-cognate transfer RNA anticodon stem-loop mismatched at the first codon position and streptomycin bound | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-16 | | Release date: | 2012-11-14 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A structural basis for streptomycin-induced misreading of the genetic code.
Nat Commun, 4, 2013
|
|
4DV0
 
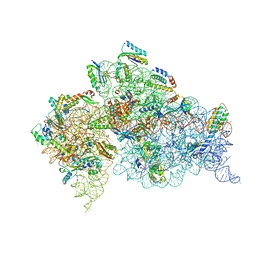 | | Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, U20G | | Descriptor: | 16S rRNA, MAGNESIUM ION, ZINC ION, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-22 | | Release date: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (3.853 Å) | | Cite: | A structural basis for streptomycin resistance
To be Published
|
|
4DYU
 
 | | The crystal structure of DNA starvation/stationary phase protection protein Dps from Yersinia pestis KIM 10 | | Descriptor: | DNA protection during starvation protein, SULFATE ION, ZINC ION | | Authors: | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-02-29 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The crystal structure of DNA starvation/stationary phase protection protein Dps from Yersinia pestis KIM 10
To be Published
|
|
4E2Q
 
 | |
4E1L
 
 | | Crystal structure of Acetoacetyl-CoA thiolase (thlA2) from Clostridium difficile | | Descriptor: | Acetoacetyl-CoA thiolase 2, IODIDE ION | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-06 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: |
|
|
4DV1
 
 | | Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, U20G, bound with streptomycin | | Descriptor: | 16S rRNA, MAGNESIUM ION, STREPTOMYCIN, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-22 | | Release date: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (3.849 Å) | | Cite: | A structural basis for streptomycin resistance
To be Published
|
|
4DZ4
 
 | |
4E2S
 
 | |
4DX5
 
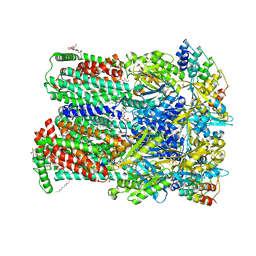 | | Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, Acriflavine resistance protein B, DARPIN, ... | | Authors: | Eicher, T, Cha, H, Seeger, M.A, Brandstaetter, L, El-Delik, J, Bohnert, J.A, Kern, W.V, Verrey, F, Gruetter, M.G, Diederichs, K, Pos, K.M. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4P3B
 
 | |
4DR1
 
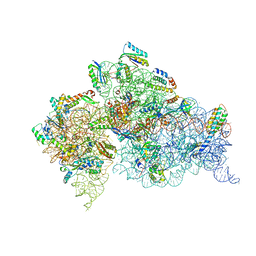 | | Crystal structure of the apo 30S ribosomal subunit from Thermus thermophilus (HB8) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-16 | | Release date: | 2012-11-14 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A structural basis for streptomycin-induced misreading of the genetic code.
Nat Commun, 4, 2013
|
|
4DV3
 
 | | Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, C912A, bound with streptomycin | | Descriptor: | 16S rRNA, MAGNESIUM ION, STREPTOMYCIN, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-22 | | Release date: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (3.547 Å) | | Cite: | A structural basis for streptomycin resistance
To be Published
|
|
4POQ
 
 | | Structure of unliganded VP1 pentamer of Human Polyomavirus 9 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ISOPROPYL ALCOHOL, ... | | Authors: | Khan, Z.M, Stehle, T. | | Deposit date: | 2014-02-26 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and glycan microarray analysis of human polyomavirus 9 VP1 identifies N-glycolyl neuraminic acid as a receptor candidate.
J.Virol., 88, 2014
|
|
4PV6
 
 | | Crystal Structure Analysis of Ard1 from Thermoplasma volcanium | | Descriptor: | ACETYL COENZYME *A, COENZYME A, N-terminal acetyltransferase complex subunit [ARD1] | | Authors: | Ma, C, Lee, S.J, Lee, B.J. | | Deposit date: | 2014-03-15 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Structure of Thermoplasma volcanium Ard1 belongs to N-acetyltransferase family member suggesting multiple ligand binding modes with acetyl coenzyme A and coenzyme A.
Biochim.Biophys.Acta, 1844, 2014
|
|
4PJ1
 
 | | Crystal structure of the human mitochondrial chaperonin symmetrical 'football' complex | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Frolow, F, Azem, A, Nisemblat, S. | | Deposit date: | 2014-05-10 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structure of the human mitochondrial chaperonin symmetrical football complex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
