4O8M
 
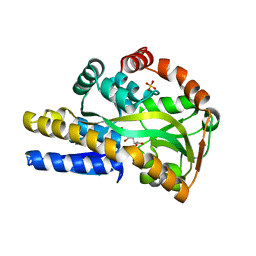 | | Crystal structure of a trap periplasmic solute binding protein actinobacillus succinogenes 130z, target EFI-510004, with bound L-galactonate | | Descriptor: | CHLORIDE ION, L-galactonic acid, SULFATE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-28 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4ELG
 
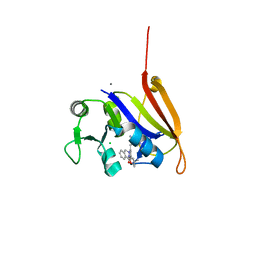 | | Structure-activity relationship guides enantiomeric preference among potent inhibitors of B. anthracis dihydrofolate reductase | | Descriptor: | (2E)-3-{5-[(2,4-diaminopyrimidin-5-yl)methyl]-2,3-dimethoxyphenyl}-1-[(1R)-1-(2-methylpropyl)phthalazin-2(1H)-yl]prop-2 -en-1-one, (2E)-3-{5-[(2,4-diaminopyrimidin-5-yl)methyl]-2,3-dimethoxyphenyl}-1-[(1S)-1-(2-methylpropyl)phthalazin-2(1H)-yl]prop-2-en-1-one, CALCIUM ION, ... | | Authors: | Bourne, C.R, Barrow, W.W. | | Deposit date: | 2012-04-10 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structure-activity relationship for enantiomers of potent inhibitors of B. anthracis dihydrofolate reductase.
Biochim.Biophys.Acta, 1834, 2013
|
|
4UTW
 
 | | Structural characterisation of NanE, ManNac6P C2 epimerase, from Clostridium perfingens | | Descriptor: | CHLORIDE ION, N-acetyl-D-glucosamine-6-phosphate, PUTATIVE N-ACETYLMANNOSAMINE-6-PHOSPHATE 2-EPIMERASE | | Authors: | Pelissier, M.C, Sebban-Kreuzer, C, Guerlesquin, F, Brannigan, J.A, Davies, G.J, Bourne, Y, Vincent, F. | | Deposit date: | 2014-07-23 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Characterization of the Clostridium Perfringens N-Acetylmannosamine-6-Phosphate 2-Epimerase Essential for the Sialic Acid Salvage Pathway
J.Biol.Chem., 289, 2014
|
|
8PDF
 
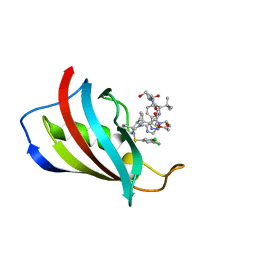 | | FKBP12 in complex with PROTAC 6a2 | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[2-[2-[2-[4-[(1~{S})-1-[(1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-ethenyl-2-oxidanylidene-3,10-diazabicyclo[4.3.1]decan-3-yl]ethyl]-1,2,3-triazol-1-yl]ethoxy]ethoxy]ethanoylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Meyners, C, Walz, M, Geiger, T.M, Hausch, F. | | Deposit date: | 2023-06-12 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery of a Potent Proteolysis Targeting Chimera Enables Targeting the Scaffolding Functions of FK506-Binding Protein 51 (FKBP51).
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
4OAY
 
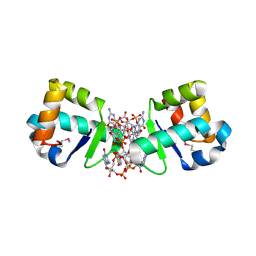 | | BldD CTD-c-di-GMP complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DNA-binding protein | | Authors: | Schumacher, M.A, Tschowri, N, Buttner, M, Brennan, R.G. | | Deposit date: | 2014-01-06 | | Release date: | 2014-11-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development.
Cell(Cambridge,Mass.), 158, 2014
|
|
7NMN
 
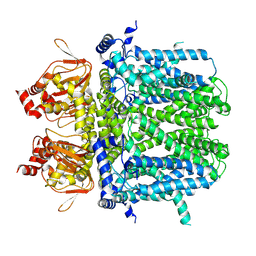 | | Rabbit HCN4 stabilised in amphipol A8-35 | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4,Rabbit HCN4 | | Authors: | Chaves-Sanjuan, A. | | Deposit date: | 2021-02-23 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Gating movements and ion permeation in HCN4 pacemaker channels.
Mol.Cell, 81, 2021
|
|
6EKN
 
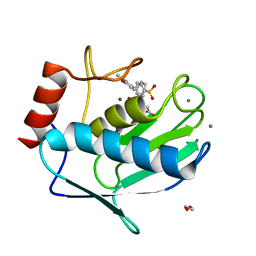 | | Crystal structure of MMP12 in complex with inhibitor BE7. | | Descriptor: | (2~{S})-2-[2-[4-(4-methoxyphenyl)phenyl]sulfonylphenyl]pentanedioic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Ciccone, L, Tepshi, L, Nuti, E, Rossello, A, Stura, E.A. | | Deposit date: | 2017-09-26 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Development of Thioaryl-Based Matrix Metalloproteinase-12 Inhibitors with Alternative Zinc-Binding Groups: Synthesis, Potentiometric, NMR, and Crystallographic Studies.
J. Med. Chem., 61, 2018
|
|
6EN0
 
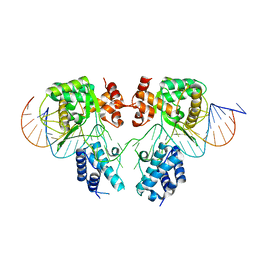 | |
6EL5
 
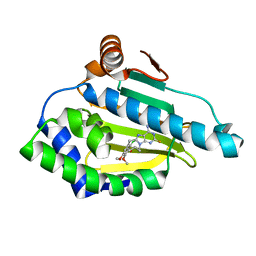 | | Estimation of relative drug-target residence times by random acceleration molecular dynamics simulation | | Descriptor: | 8-(2-CHLORO-3,4,5-TRIMETHOXY-BENZYL)-2-FLUORO-9-PENT-4-YLNYL-9H-PURIN-6-YLAMINE, Heat shock protein HSP 90-alpha | | Authors: | Musil, D, Lehmann, M, Eggenweiler, H.-M. | | Deposit date: | 2017-09-27 | | Release date: | 2018-05-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Estimation of Drug-Target Residence Times by tau-Random Acceleration Molecular Dynamics Simulations.
J Chem Theory Comput, 14, 2018
|
|
6ELJ
 
 | | FAB Fragment. AbVance: Increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery | | Descriptor: | fAB heavy chain, fAB light chain | | Authors: | Benz, J, Weigand, S, Dengl, S, Schlothauer, T, Auer, J, Ehler, A, Kettenberger, H, Lorenz, S, Hirschheydt, T, Georges, G. | | Deposit date: | 2017-09-29 | | Release date: | 2017-11-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | AbVance: increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery
To Be Published
|
|
6EOP
 
 | | DPP8 - SLRFLYEG, space group 20 | | Descriptor: | CALCIUM ION, CITRATE ANION, Dipeptidyl peptidase 8, ... | | Authors: | Ross, B.R, Huber, R. | | Deposit date: | 2017-10-10 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures and mechanism of dipeptidyl peptidases 8 and 9, important players in cellular homeostasis and cancer.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4WDG
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation V321A, complexed with 2',5'-ADP | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, ADENOSINE-2'-5'-DIPHOSPHATE | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-08 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
4EV9
 
 | |
4EVP
 
 | |
4OGB
 
 | | Crystal structure of the catalytic domain of PDE4D2 with compound 2 | | Descriptor: | (2R)-8-(3,4-dimethoxyphenyl)-6-methyl-2-(tetrahydro-2H-pyran-4-yl)-2H-chromen-4-ol, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Feil, S.C, Parker, M.W. | | Deposit date: | 2014-01-15 | | Release date: | 2015-01-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.032 Å) | | Cite: | The PDE inhibition profile of LY294002 and tetrahydropyranyl analogues reveals a chromone motif for the development of PDE inhibitors
To be Published
|
|
6EO8
 
 | | Crystal structure of thrombin in complex with a novel glucose-conjugated potent inhibitor | | Descriptor: | DIMETHYL SULFOXIDE, Hirudin variant-2, N-(2-{[5-(5-chlorothiophen-2-yl)-1,2-oxazol-3-yl]methoxy}-6-[3-(beta-D-glucopyranosyloxy)propoxy]phenyl)-1-(propan-2-yl)piperidine-4-carboxamide, ... | | Authors: | Belviso, B.D, Caliandro, R, Aresta, B.M, De Candia, M, Altomare, C.D. | | Deposit date: | 2017-10-09 | | Release date: | 2017-12-13 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | How a beta-D-glucoside side chain enhances binding affinity to thrombin of inhibitors bearing 2-chlorothiophene as P1 moiety: crystallography, fragment deconstruction study, and evaluation of antithrombotic properties.
J. Med. Chem., 57, 2014
|
|
6EOT
 
 | | DPP8 - SLRFLYEG, space group 19 | | Descriptor: | Dipeptidyl peptidase 8, SER-LEU-ARG-PHE-LEU-TYR-GLU-GLY | | Authors: | Ross, B.R, Huber, R. | | Deposit date: | 2017-10-10 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structures and mechanism of dipeptidyl peptidases 8 and 9, important players in cellular homeostasis and cancer.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6E5A
 
 | | PPARg in complex with compound 4b | | Descriptor: | (5Z)-5-({4-[(prop-2-yn-1-yl)oxy]phenyl}methylidene)-2-sulfanylidene-1,3-thiazolidin-4-one, Peroxisome proliferator-activated receptor gamma | | Authors: | Bruning, J.B, Chua, B.S.K. | | Deposit date: | 2018-07-19 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Shooting three inflammatory targets with a single bullet: Novel multi-targeting anti-inflammatory glitazones.
Eur J Med Chem, 167, 2019
|
|
6E5Z
 
 | |
6EAH
 
 | |
7N1F
 
 | | SARS-CoV-2 YLQ peptide-specific TCR pYLQ7 binds to YLQ-HLA-A2 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, A-2 alpha chain, ... | | Authors: | Wu, D, Mariuzza, R.A. | | Deposit date: | 2021-05-27 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Structural assessment of HLA-A2-restricted SARS-CoV-2 spike epitopes recognized by public and private T-cell receptors.
Nat Commun, 13, 2022
|
|
4OH2
 
 | | Crystal Structure of Cu/Zn Superoxide Dismutase I149T | | Descriptor: | COPPER (II) ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Crane, B.R, Merz, G.E. | | Deposit date: | 2014-01-16 | | Release date: | 2014-10-15 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.384 Å) | | Cite: | Copper-Based Pulsed Dipolar ESR Spectroscopy as a Probe of Protein Conformation Linked to Disease States.
Biophys.J., 107, 2014
|
|
7N65
 
 | | Complex structure of HIV superinfection Fab QA013.2 and BG505.SOSIP.664 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp41, ... | | Authors: | Mangala Prasad, V, Shipley, M.M, Overbaugh, J.M, Lee, K.K. | | Deposit date: | 2021-06-07 | | Release date: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Functional development of a V3/glycan-specific broadly neutralizing antibody isolated from a case of HIV superinfection.
Elife, 10, 2021
|
|
4ES6
 
 | |
4OE7
 
 | | Crystal structure of YagE, a KDG aldolase protein, in complex with aldol condensed product of pyruvate and glyoxal | | Descriptor: | (4R)-4-hydroxy-2,5-dioxopentanoic acid, (4S)-4-hydroxy-2,5-dioxopentanoic acid, 1,2-ETHANEDIOL, ... | | Authors: | Manoj Kumar, P, Baskar, V, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2014-01-12 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of YagE, a KDG aldolase protein, in complex with aldol condensed product of pyruvate and glyoxal
To be Published
|
|
