7R7Z
 
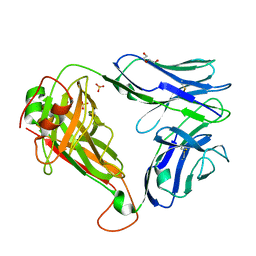 | | Crystal structure of QW9-HLA-B*5301 specific T Cell Receptor, C3 | | Descriptor: | Alpha chain of C3 TCR, Beta chain of C3 TCR, HEXAETHYLENE GLYCOL, ... | | Authors: | Li, X.L, Tan, K.M, Xu, S.T, Ng, R, Walker, B.D, Wang, J.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.286 Å) | | Cite: | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
7ABH
 
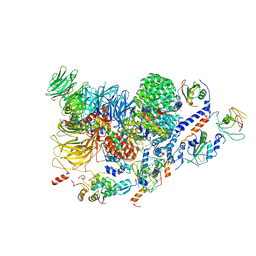 | | Human pre-Bact-2 spliceosome (SF3b/U2 snRNP portion) | | Descriptor: | Cell division cycle 5-like protein, DNA/RNA-binding protein KIN17, MINX M3 pre-mRNA, ... | | Authors: | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | Deposit date: | 2020-09-07 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
7R7V
 
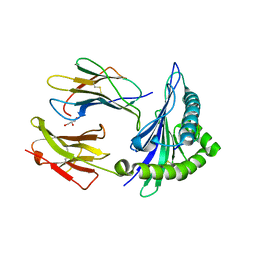 | | Crystal structure of HLA-B*5301 complex with an HIV-1 Gag-derived epitope QW9 | | Descriptor: | Beta-2-microglobulin, GLN-ALA-SER-GLN-GLU-VAL-LYS-ASN-TRP, GLYCEROL, ... | | Authors: | Li, X.L, Tan, K.M, Walker, B.D, Wang, J.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
7R7W
 
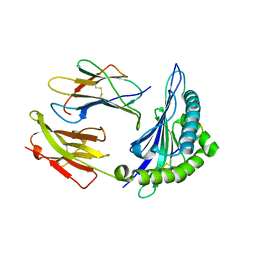 | | Crystal structure of HLA-B*5301 complex with an HIV-1 Gag-derived epitope QW9 S3T variant | | Descriptor: | Beta-2-microglobulin, GLN-ALA-THR-GLN-GLU-VAL-LYS-ASN-TRP, MHC class I antigen | | Authors: | Li, X.L, Tan, K.M, Walker, B.D, Wang, J.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
7R80
 
 | | Crystal structure of C3 TCR complex with QW9-bound HLA-B*5301 | | Descriptor: | Alpha chain of C3 TCR, Beta Chain of C3 TCR, Beta-2-microglobulin, ... | | Authors: | Li, X.L, Tan, K.M, Walker, B.D, Wang, J.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
6QTP
 
 | | 2.37A structure of gepotidacin with S.aureus DNA gyrase and uncleaved DNA | | Descriptor: | (3~{R})-3-[[4-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)piperidin-1-yl]methyl]-1,4,7-triazatricyclo[6.3.1.0^{4,12}]dodeca-6,8(12),9-triene-5,11-dione, DNA (5'-D(*GP*AP*GP*CP*GP*TP*AP*CP*AP*GP*CP*TP*GP*TP*AP*CP*GP*CP*TP*T)-3'), DNA gyrase subunit A, ... | | Authors: | Bax, B.D. | | Deposit date: | 2019-02-25 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Mechanistic and Structural Basis for the Actions of the Antibacterial Gepotidacin against Staphylococcus aureus Gyrase.
Acs Infect Dis., 5, 2019
|
|
8FCX
 
 | |
6HX3
 
 | | PDX1.2/PDX1.3 complex | | Descriptor: | Pyridoxal 5'-phosphate synthase subunit PDX1.3, Pyridoxal 5'-phosphate synthase-like subunit PDX1.2, SULFATE ION | | Authors: | Robinson, G.C, Kaufmann, M, Roux, C, Martinez-Font, J, Hothorn, M, Thore, S, Fitzpatrick, T.B. | | Deposit date: | 2018-10-15 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the pseudoenzyme PDX1.2 in complex with its cognate enzyme PDX1.3: a total eclipse.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8FCW
 
 | |
8FCV
 
 | |
8OKX
 
 | | Structure of cGAS in complex with SPSB3-ELOBC | | Descriptor: | Cyclic GMP-AMP synthase, Elongin-B, Elongin-C, ... | | Authors: | Xu, P.B, Ablasser, A. | | Deposit date: | 2023-03-29 | | Release date: | 2024-02-14 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | The CRL5-SPSB3 ubiquitin ligase targets nuclear cGAS for degradation.
Nature, 627, 2024
|
|
8UT7
 
 | | CryoEM structure of A/Perth/16/2009 H3 in complex with polyclonal Fab from mice immunized with H3 stem nanoparticles-28 days post immunization | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, H3D28 pFab HC Fv_polyA, ... | | Authors: | Huang, J, Han, J, Ward, A.B. | | Deposit date: | 2023-10-30 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Eliciting a single amino acid change by vaccination generates antibody protection against group 1 and group 2 influenza A viruses.
Immunity, 57, 2024
|
|
6HYE
 
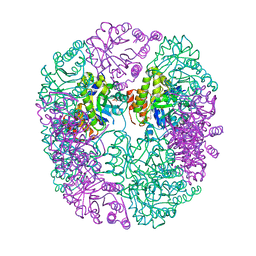 | | PDX1.2/PDX1.3 complex (PDX1.3:K97A) | | Descriptor: | Pyridoxal 5'-phosphate synthase subunit PDX1.3, Pyridoxal 5'-phosphate synthase-like subunit PDX1.2, SULFATE ION | | Authors: | Robinson, G.C, Kaufmann, M, Roux, C, Martinez-Font, J, Hothorn, M, Thore, S, Fitzpatrick, T.B. | | Deposit date: | 2018-10-20 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal structure of the pseudoenzyme PDX1.2 in complex with its cognate enzyme PDX1.3: a total eclipse.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8D6C
 
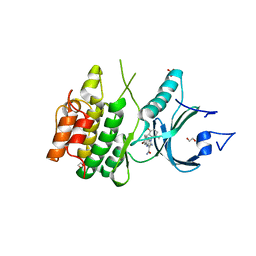 | | Crystal Structure of Human Myt1 Kinase domain Bounded with compound 28 | | Descriptor: | (1P)-2-amino-6-bromo-1-(3-hydroxy-2,6-dimethylphenyl)-1H-pyrrolo[2,3-b]quinoxaline-3-carboxamide, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Pau, V.P.T, Mao, D.Y.L, Mader, P, Orlicky, S, Sicheri, F. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of an Orally Bioavailable and Selective PKMYT1 Inhibitor, RP-6306.
J.Med.Chem., 65, 2022
|
|
8T2H
 
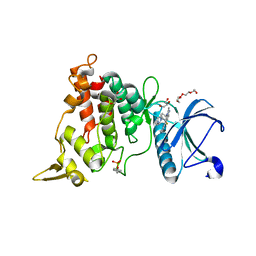 | | DYRK1A complex with DYR530 | | Descriptor: | (4P)-4-{(3M)-3-[3-fluoro-4-(4-methylpiperazin-1-yl)phenyl]-2-methyl-3H-imidazo[4,5-b]pyridin-5-yl}pyridin-2-amine, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, GLYCEROL, ... | | Authors: | Montfort, W.R, Basantes, L.E. | | Deposit date: | 2023-06-06 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of DYR684, a Potent, Selective, Metabolically Stable, DYRK1A/B PROTAC utilizing a Novel Cereblon Molecular Glue
To Be Published
|
|
7A02
 
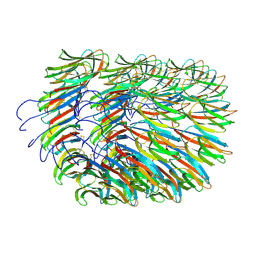 | | Bacillus endospore appendages form a novel family of disulfide-linked pili | | Descriptor: | DUF3992 domain-containing protein | | Authors: | Pradhan, B, Liedtke, J, Sleutel, M, Lindback, T, Llarena, A.K, Brynildsrud, O, Aspholm, M, Remaut, H. | | Deposit date: | 2020-08-06 | | Release date: | 2021-04-28 | | Last modified: | 2021-09-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Endospore Appendages: a novel pilus superfamily from the endospores of pathogenic Bacilli.
Embo J., 40, 2021
|
|
5NAI
 
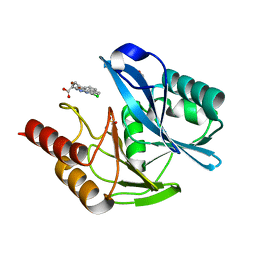 | | mono-Zinc VIM-5 metallo-beta-lactamase in complex with (1-chloro-4-hydroxyisoquinoline-3-carbonyl)-D-tryptophan (Compound 1) | | Descriptor: | (2~{R})-2-[(1-chloranyl-4-oxidanyl-isoquinolin-3-yl)carbonylamino]-3-(1~{H}-indol-3-yl)propanoic acid, Class B metallo-beta-lactamase, FLUORIDE ION, ... | | Authors: | Li, G.-B, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2017-02-28 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystallographic analyses of isoquinoline complexes reveal a new mode of metallo-beta-lactamase inhibition.
Chem. Commun. (Camb.), 53, 2017
|
|
7MBO
 
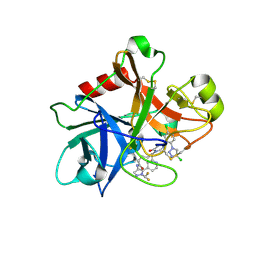 | | FACTOR XIA (PICHIA PASTORIS; C500S [C122S]) IN COMPLEX WITH THE INHIBITOR Milvexian (BMS-986177), IUPAC NAME:(6R,10S)-10-{4-[5-chloro-2-(4-chloro-1H-1,2,3-triazol-1-yl)phenyl]-6- oxopyrimidin-1(6H)-yl}-1-(difluoromethyl)-6-methyl-1,4,7,8,9,10-hexahydro-15,11- (metheno)pyrazolo[4,3-b][1,7]diazacyclotetradecin-5(6H)-one | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XIa light chain, Milvexian | | Authors: | Sheriff, S. | | Deposit date: | 2021-04-01 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.924 Å) | | Cite: | Discovery of Milvexian, a High-Affinity, Orally Bioavailable Inhibitor of Factor XIa in Clinical Studies for Antithrombotic Therapy.
J.Med.Chem., 65, 2022
|
|
6ZQF
 
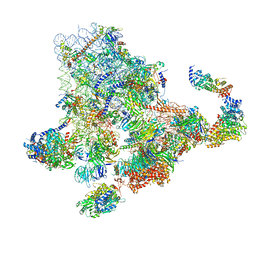 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state Dis-B (Poly-Ala) | | Descriptor: | 18S rRNA, 40S ribosomal protein S1-A, 40S ribosomal protein S11-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
6UL4
 
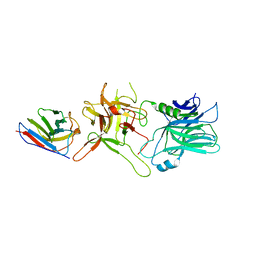 | |
8UT5
 
 | | CryoEM structure of A/Michigan/45/2015 H1 in complex with flu HA central stem VH1-18 antibody UCA6_N55T | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain, ... | | Authors: | Huang, J, Han, J, Ward, A.B. | | Deposit date: | 2023-10-30 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Eliciting a single amino acid change by vaccination generates antibody protection against group 1 and group 2 influenza A viruses.
Immunity, 57, 2024
|
|
8UT4
 
 | | CryoEM structure of A/Michigan/45/2015 H1 in complex with flu HA central stem VH1-18 antibody 09-1B12 | | Descriptor: | 09-1B12 HC Fv, 09-1B12 LC Fv, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Huang, J, Han, J, Ward, A.B. | | Deposit date: | 2023-10-30 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Eliciting a single amino acid change by vaccination generates antibody protection against group 1 and group 2 influenza A viruses.
Immunity, 57, 2024
|
|
4X0C
 
 | |
8VXE
 
 | | Structure of p38 alpha (Mitogen-activated protein kinase 14) complexed with inhibitor 6 | | Descriptor: | (4M)-4-[3-(4-fluorophenyl)-1-methyl-1H-pyrazol-4-yl]-1H-pyrrolo[2,3-b]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Blaesse, M, Steinbacher, S, Shaffer, P.L, Sharma, S, Thompson, A.A. | | Deposit date: | 2024-02-04 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Optimization of Selective and Brain Penetrant CK1 delta Inhibitors for the Treatment of Circadian Disruptions.
Acs Med.Chem.Lett., 15, 2024
|
|
7XT6
 
 | | Structure of a membrane protein M3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, B-cell antigen receptor complex-associated protein alpha chain, ... | | Authors: | Ma, X, Zhu, Y, Chen, Y, Huang, Z. | | Deposit date: | 2022-05-16 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Cryo-EM structures of two human B cell receptor isotypes.
Science, 377, 2022
|
|
