2JUM
 
 | | ThrA3-DKP-insulin | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Huang, K, Chan, S, Hua, Q, Chu, Y, Wang, R, Klaproth, B, Jia, W, Whittaker, J, De Meyts, P, Nakagawa, S.H, Steiner, D.F, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2007-08-31 | | Release date: | 2007-10-16 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | The A-chain of Insulin Contacts the Insert Domain of the Insulin Receptor: PHOTO-CROSS-LINKING AND MUTAGENESIS OF A DIABETES-RELATED CREVICE.
J.Biol.Chem., 282, 2007
|
|
3PQS
 
 | |
3TTQ
 
 | | Crystal structure of Leuconostoc mesenteroides NRRL B-1299 N-terminally truncated dextransucrase DSR-E in orthorhombic apo-form at 1.9 angstrom resolution | | Descriptor: | CALCIUM ION, Dextransucrase, GLYCEROL, ... | | Authors: | Brison, Y, Pijning, T, Fabre, E, Mourey, L, Morel, S, Potocki-Veronese, G, Monsan, P, Remaud-Simeon, M, Dijkstra, B.W, Tranier, S. | | Deposit date: | 2011-09-15 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional and structural characterization of alpha-(1-2) branching sucrase derived from DSR-E glucansucrase
J.Biol.Chem., 287, 2012
|
|
2JUV
 
 | | AbaA3-DKP-insulin | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Huang, K, Chan, S, Hua, Q, Chu, Y, Wang, R, Klaproth, B, Jia, W, Whittaker, J, De Meyts, P, Nakagawa, S.H, Steiner, D.F, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2007-09-05 | | Release date: | 2007-10-16 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | The A-chain of Insulin Contacts the Insert Domain of the Insulin Receptor: PHOTO-CROSS-LINKING AND MUTAGENESIS OF A DIABETES-RELATED CREVICE.
J.Biol.Chem., 282, 2007
|
|
1DV9
 
 | | STRUCTURAL CHANGES ACCOMPANYING PH-INDUCED DISSOCIATION OF THE B-LACTOGLOBULIN DIMER | | Descriptor: | BETA-LACTOGLOBULIN | | Authors: | Uhrinova, S, Smith, M.H, Jameson, G.B, Uhrin, D, Sawyer, L, Barlow, P.N. | | Deposit date: | 2000-01-20 | | Release date: | 2000-02-09 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structural changes accompanying pH-induced dissociation of the beta-lactoglobulin dimer.
Biochemistry, 39, 2000
|
|
3Q6W
 
 | | Structure of dually-phosphorylated MET receptor kinase in complex with an MK-2461 analog with specificity for the activated receptor | | Descriptor: | 3-{5-oxo-3-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]-5H-benzo[4,5]cyclohepta[1,2-b]pyridin-7-yl}-N-(pyridin-2-ylmethyl)propanamide, Hepatocyte growth factor receptor | | Authors: | Soisson, S.M, Rickert, K.W, Patel, S.B, Lumb, K.J. | | Deposit date: | 2011-01-03 | | Release date: | 2011-01-19 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for selective small molecule kinase inhibition of activated c-Met.
J.Biol.Chem., 286, 2011
|
|
2QMT
 
 | | Crystal Polymorphism of Protein GB1 Examined by Solid-state NMR and X-ray Diffraction | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ISOPROPYL ALCOHOL, Immunoglobulin G-binding protein G, ... | | Authors: | Frericks Schmidt, H.L, Sperling, L.J, Gao, Y.G, Wylie, B.J, Boettcher, J.M, Wilson, S.R, Rienstra, C.M. | | Deposit date: | 2007-07-16 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Crystal Polymorphism of Protein GB1 Examined by Solid-State NMR Spectroscopy and X-ray Diffraction.
J.Phys.Chem.B, 111, 2007
|
|
1CIM
 
 | | THE POSITIONS OF HIS-64 AND A BOUND WATER IN HUMAN CARBONIC ANHYDRASE II UPON BINDING THREE STRUCTURALLY RELATED INHIBITORS | | Descriptor: | (4S-TRANS)-4-(AMINO)-5,6-DIHYDRO-6-METHYL-4H-THIENO (2,3-B)THIOPYRAN-2-SULFONAMIDE-7,7-DIOXIDE, CARBONIC ANHYDRASE II, METHYL MERCURY ION, ... | | Authors: | Smith, G.M, Alexander, R.S, Christianson, D.W, Mckeever, B.M, Ponticello, G.S, Springer, J.P, Randall, W.C, Baldwin, J.J, Habecker, C.N. | | Deposit date: | 1993-10-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Positions of His-64 and a bound water in human carbonic anhydrase II upon binding three structurally related inhibitors.
Protein Sci., 3, 1994
|
|
3CZ4
 
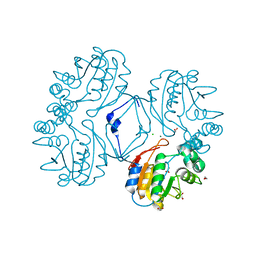 | | Native AphA class B acid phosphatase/phosphotransferase from E. coli | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Leone, R, Cappelletti, E, Benvenuti, M, Lentini, G, Thaller, M.C, Mangani, S. | | Deposit date: | 2008-04-28 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the catalytic mechanism of the bacterial class B phosphatase AphA belonging to the DDDD superfamily of phosphohydrolases.
J.Mol.Biol., 384, 2008
|
|
1WG3
 
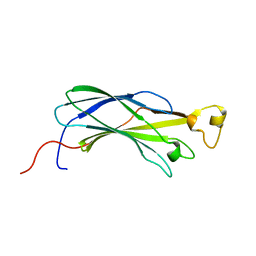 | | Structural analysis of yeast nucleosome-assembly factor CIA1p | | Descriptor: | Anti-silencing protein 1 | | Authors: | Padmanabhan, B, Kataoka, K, Umehara, T, Adachi, N, Yokoyama, S, Horikoshi, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-27 | | Release date: | 2005-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Similarity between Histone Chaperone Cia1p/Asf1p and DNA-Binding Protein NF-{kappa}B
J.Biochem.(Tokyo), 138, 2005
|
|
1JMP
 
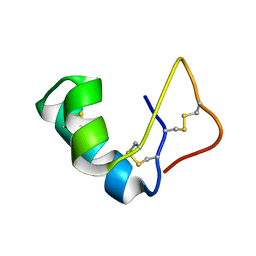 | | Solution Structure of the Viscotoxin B | | Descriptor: | viscotoxin B | | Authors: | Coulon, A, Mosbah, A, Bernard, C, Rouge, P, Urech, K, Darbon, H. | | Deposit date: | 2001-07-19 | | Release date: | 2003-11-11 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Comparative membrane interaction study of viscotoxins A3, A2 and B from mistletoe (Viscum album) and connections with their structures
Biochem.J., 374, 2003
|
|
1K8H
 
 | |
2IXY
 
 | | Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal | | Descriptor: | 5'-R(*GP*GP*CP*CP*UP*CP*CP*AP*AP*GP *CP*UP*GP*UP*GP*CP*CP*UP*UP*GP*GP*GP*UP*GP*GP*CP*C)-3' | | Authors: | Flodell, S, Petersen, M, Girard, F, Zdunek, J, Kidd-Ljunggren, K, Schleucher, J, Wijmenga, S.S. | | Deposit date: | 2006-07-11 | | Release date: | 2006-09-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal.
Nucleic Acids Res., 34, 2006
|
|
2F1N
 
 | |
3CD8
 
 | | X-ray Structure of c-Met with triazolopyridazine Inhibitor. | | Descriptor: | 7-methoxy-4-[(6-phenyl[1,2,4]triazolo[4,3-b]pyridazin-3-yl)methoxy]quinoline, Hepatocyte growth factor receptor | | Authors: | Bellon, S.F, Albrecht, B.K, Harmange, J.-C, Bauer, D, Choquette, D, Dussault, I. | | Deposit date: | 2008-02-26 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and Optimization of Triazolopyridazines as Potent and Selective Inhibitors of the c-Met Kinase.
J.Med.Chem., 51, 2008
|
|
3R7O
 
 | | Structure of dually phosphorylated c-MET receptor kinase in complex with an MK-2461 analog | | Descriptor: | Hepatocyte growth factor receptor, N-[(2R)-1,4-dioxan-2-ylmethyl]-N-methyl-N'-{5-oxo-3-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]-5H-benzo[4,5]cyclohepta[1,2-b]pyridin-7-yl}sulfuric diamide | | Authors: | Soisson, S.M, Rickert, K, Patel, S.B, Munshi, S, Lumb, K.J. | | Deposit date: | 2011-03-22 | | Release date: | 2012-02-01 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for selective small molecule kinase inhibition of activated c-Met.
J.Biol.Chem., 286, 2011
|
|
3S4J
 
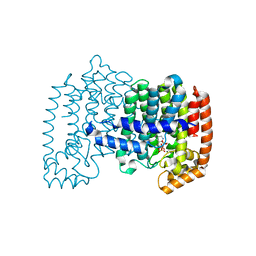 | | Human FDPS Synthase in Complex with a Rigid Analog of Risedronate | | Descriptor: | 6,7-dihydro-5H-cyclopenta[b]pyridine-6,6-diylbis(phosphonic acid), Farnesyl pyrophosphate synthase, MAGNESIUM ION, ... | | Authors: | Evdokimov, A.G, Barnett, B.L, Ebetino, F.H, Pokross, M. | | Deposit date: | 2011-05-19 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Human FDPS Synthase in Complex with a Rigid Analog of Risedronate
To be Published
|
|
3TB7
 
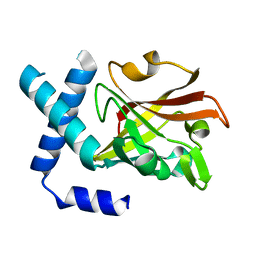 | |
2MVG
 
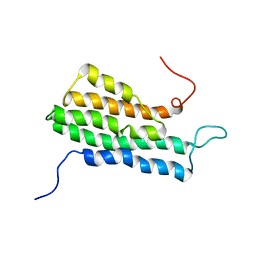 | |
3U89
 
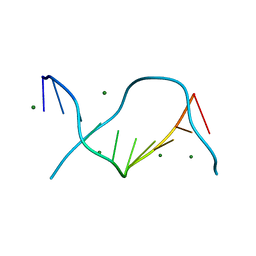 | | Crystal structure of one turn of g/c rich b-dna revisited | | Descriptor: | 5'-D(*CP*CP*AP*GP*GP*CP*CP*TP*GP*G) -3', MAGNESIUM ION | | Authors: | Maehigashi, T, Woods, K.K, Moulaei, T, Komeda, S, Williams, L.D. | | Deposit date: | 2011-10-16 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | B-DNA structure is intrinsically polymorphic: even at the level of base pair positions.
Nucleic Acids Res., 40, 2012
|
|
2BY4
 
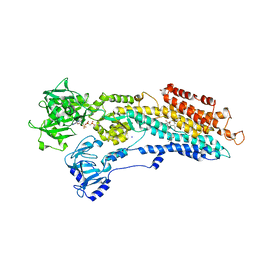 | | SR Ca(2+)-ATPase in the HnE2 state complexed with the thapsigargin derivative Boc-12ADT. | | Descriptor: | (3S,3AR,4S,6S,6AR,7S,8S,9BS)-6-(ACETYLOXY)-3,3A-DIHYDROXY-3,6,9-TRIMETHYL-8-{[(2Z)-2-METHYLBUT-2-ENOYL]OXY}-7-(OCTANOYLOXY)-2-OXO-2,3,3A,4,5,6,6A,7,8,9B-DECAHYDROAZULENO[4,5-B]FURAN-4-YL 12-[(TERT-BUTOXYCARBONYL)AMINO]DODECANOATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Jensen, A.L, Moller, J.V, Soehoel, H, Denmeade, S.R, Isaacs, J.T, Olsen, C.E, Christensen, S.B, Nissen, P. | | Deposit date: | 2005-07-28 | | Release date: | 2006-04-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Natural Products as Starting Materials for Development of Second-Generation Serca Inhibitors Targeted Towards Prostate Cancer Cells
Bioorg.Med.Chem., 14, 2006
|
|
2GHG
 
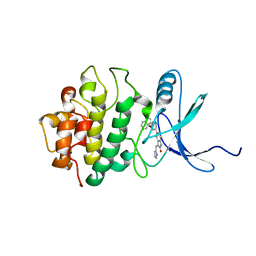 | | h-CHK1 complexed with A431994 | | Descriptor: | 5-{5-[(S)-2-AMINO-3-(1H-INDOL-3-YL)-PROPOXYL]-PYRIDIN-3-YL}-3-[1-(1H-PYRROL-2-YL)-METH-(Z)-YLIDENE]-1,3-DIHYDRO-INDOL-2-ONE, Serine/threonine-protein kinase Chk1 | | Authors: | Park, C. | | Deposit date: | 2006-03-27 | | Release date: | 2007-03-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Discovery and SAR of oxindole-pyridine-based protein kinase B/Akt inhibitors for treating cancers.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1X0O
 
 | | human ARNT C-terminal PAS domain | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator | | Authors: | Card, P.B, Erbel, P.J, Gardner, K.H. | | Deposit date: | 2005-03-25 | | Release date: | 2005-10-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of ARNT PAS-B Dimerization: Use of a Common Beta-sheet Interface for Hetero- and Homodimerization.
J.Mol.Biol., 353, 2005
|
|
5WK5
 
 | |
3V57
 
 | |
