1F10
 
 | |
2X3A
 
 | | AsaP1 inactive mutant E294Q, an extracellular toxic zinc metalloendopeptidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Bogdanovic, X, Palm, G.J, Singh, R.K, Hinrichs, W. | | Deposit date: | 2010-01-22 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Evidence of Intramolecular Propeptide Inhibition of the Aspzincin Metalloendopeptidase Asap1.
FEBS Lett., 590, 2016
|
|
1A9Z
 
 | |
4HMN
 
 | | Crystal structure of human 17beta-hydroxysteroid dehydrogenase type 5 in complex with (4-(4-Chlorophenyl)piperazin-1-yl)(morpholino)methanone (24) | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Turnbull, A.P, Flanagan, J.U, Atwell, G.J, Heinrich, D.M, Jamieson, S.M.F, Brooke, D.G, Silva, S, Rigoreau, L.J.M, Trivier, E, Soudy, C, Samlal, S.S, Owen, P.J, Schroeder, E, Raynham, T, Denny, W.A. | | Deposit date: | 2012-10-18 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Morpholylureas are a new class of potent and selective inhibitors of the type 5 17-beta-hydroxysteroid dehydrogenase (AKR1C3).
Bioorg.Med.Chem., 22, 2014
|
|
1A5L
 
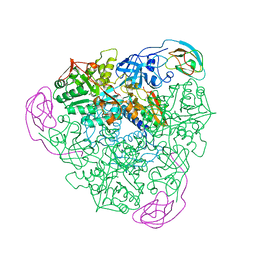 | | K217C VARIANT OF KLEBSIELLA AEROGENES UREASE | | Descriptor: | UREASE (ALPHA SUBUNIT), UREASE (BETA SUBUNIT), UREASE (GAMMA SUBUNIT) | | Authors: | Pearson, M.A, Schaller, R.A, Michel, L.O, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 1998-02-17 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chemical rescue of Klebsiella aerogenes urease variants lacking the carbamylated-lysine nickel ligand.
Biochemistry, 37, 1998
|
|
4HMT
 
 | | Crystal structure of PhzG from Pseudomonas fluorescens 2-79 in complex with hexahydrophenazine-1,6-dicarboxylic acid | | Descriptor: | (1R,5aS,6R)-1,2,5,5a,6,7-hexahydrophenazine-1,6-dicarboxylic acid, FLAVIN MONONUCLEOTIDE, Phenazine biosynthesis protein phzG, ... | | Authors: | Xu, N.N, Ahuja, E.G, Blankenfeldt, W. | | Deposit date: | 2012-10-18 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Trapped intermediates in crystals of the FMN-dependent oxidase PhzG provide insight into the final steps of phenazine biosynthesis
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1PQ1
 
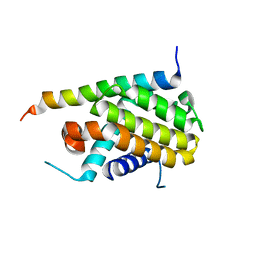 | | Crystal structure of Bcl-xl/Bim | | Descriptor: | Apoptosis regulator Bcl-X, BCL2-like protein 11 | | Authors: | Liu, X, Dai, S, Zhu, Y, Marrack, P, Kappler, J.W. | | Deposit date: | 2003-06-17 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structure of a Bcl-xl/Bim fragment complex: Implications for Bim function
Immunity, 19, 2003
|
|
3VTR
 
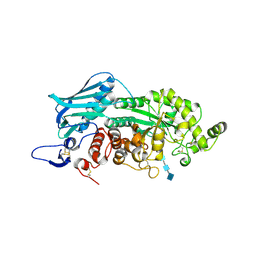 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 E328A complexed with TMG-chitotriomycin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-2-(trimethylammonio)-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetylglucosaminidase | | Authors: | Liu, T, Zhou, Y, Chen, L, Chen, W, Liu, L, Shen, X, Yang, Q. | | Deposit date: | 2012-06-02 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into cellulolytic and chitinolytic enzymes revealing crucial residues of insect beta-N-acetyl-D-hexosaminidase
Plos One, 7, 2012
|
|
1A9Y
 
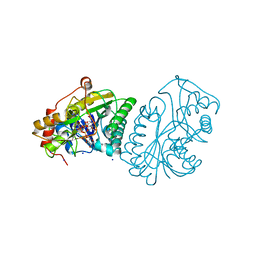 | |
1F3H
 
 | | X-RAY CRYSTAL STRUCTURE OF THE HUMAN ANTI-APOPTOTIC PROTEIN SURVIVIN | | Descriptor: | SULFATE ION, SURVIVIN, ZINC ION | | Authors: | Verdecia, M.A, Huang, H, Dutil, E, Hunter, T, Noel, J.P. | | Deposit date: | 2000-06-03 | | Release date: | 2000-12-06 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure of the human anti-apoptotic protein survivin reveals a dimeric arrangement.
Nat.Struct.Biol., 7, 2000
|
|
2ZAX
 
 | | Crystal Structure of Ferric Cytochrome P450cam | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, Cytochrome P450-cam, ... | | Authors: | Sakurai, K, Shimada, H, Harada, K, Hayashi, T, Tsukihara, T. | | Deposit date: | 2007-10-11 | | Release date: | 2008-01-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evaluation of the functional role of the heme-6-propionate side chain in cytochrome P450cam
J.Am.Chem.Soc., 130, 2008
|
|
2ZB8
 
 | | Crystal structure of human 15-ketoprostaglandin delta-13-reductase in complex with NADP and indomethacin | | Descriptor: | INDOMETHACIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Prostaglandin reductase 2, ... | | Authors: | Wu, Y.H, Wang, A.H.J, Ko, T.P, Guo, R.T, Hu, S.M, Chuang, L.M. | | Deposit date: | 2007-10-16 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of human prostaglandin reductase PTGR2.
Structure, 16, 2008
|
|
1TXG
 
 | | Structure of glycerol-3-phosphate dehydrogenase from Archaeoglobus fulgidus | | Descriptor: | AMMONIUM ION, GLYCEROL, SULFATE ION, ... | | Authors: | Sakasegawa, S, Hagemeier, C.H, Thauer, R.K, Essen, L.O, Shima, S. | | Deposit date: | 2004-07-05 | | Release date: | 2004-12-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional analysis of the gpsA gene product of Archaeoglobus fulgidus: A glycerol-3-phosphate dehydrogenase with an unusual NADP+ preference
Protein Sci., 13, 2004
|
|
1AJA
 
 | |
2ZBN
 
 | |
6T40
 
 | | Bovine enterovirus F3 in complex with a Cysteinylglycine dipeptide | | Descriptor: | CHLORIDE ION, CYSTEINE, GLYCEROL, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Walter, T.S, Fry, E.E, Stuart, D.I. | | Deposit date: | 2019-10-11 | | Release date: | 2020-08-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Glutathione facilitates enterovirus assembly by binding at a druggable pocket.
Commun Biol, 3, 2020
|
|
1TY9
 
 | | X-RAY CRYSTAL STRUCTURE OF PHZG FROM PSEUDOMONAS FLUORESCENS | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phenazine biosynthesis protein phzG, SULFATE ION | | Authors: | Parsons, J.F, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2004-07-07 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the phenazine biosynthesis enzyme PhzG.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3VXD
 
 | | Crystal structure of unsaturated glucuronyl hydrolase mutant D115N from Streptcoccus agalactiae | | Descriptor: | Putative uncharacterized protein gbs1889, SULFATE ION | | Authors: | Nakamichi, Y, Maruyama, Y, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2012-09-11 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of unsaturated glucuronyl hydrolase mutant D115N from Streptcoccus agalactiae
To be Published
|
|
2ZEZ
 
 | | Family 16 Carbohydrate Binding Module-2 | | Descriptor: | CALCIUM ION, S-layer associated multidomain endoglucanase | | Authors: | Bae, B, Nair, S.K. | | Deposit date: | 2007-12-18 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis for the Selectivity and Specificity of Ligand Recognition by the Family 16 Carbohydrate-binding Modules from Thermoanaerobacterium polysaccharolyticum ManA
J.Biol.Chem., 283, 2008
|
|
6D6X
 
 | | HSP40 co-chaperone Sis1 J-domain | | Descriptor: | Type II HSP40 co-chaperone | | Authors: | Pinheiro, G.M.S, Amorim, G.C, Iqbal, A, Ramos, C.H.I, Almeida, F.C.L. | | Deposit date: | 2018-04-23 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR investigation on the structure and function of the isolated J-domain from Sis1: Evidence of transient inter-domain interactions in the full-length protein.
Arch.Biochem.Biophys., 669, 2019
|
|
3W1U
 
 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-3-111 | | Descriptor: | 5-[2-(4-methylphenyl)ethyl]-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase (fumarate), ... | | Authors: | Inaoka, D.K, Iida, M, Tabuchi, T, Lee, N, Matsuoka, S, Shiba, T, Sakamoto, K, Suzuki, S, Balogun, E.O, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2012-11-21 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-3-111
To be Published
|
|
3VZ0
 
 | | Structural insights into cofactor and substrate selection by Gox0499 | | Descriptor: | NONAETHYLENE GLYCOL, Putative NAD-dependent aldehyde dehydrogenase | | Authors: | Yuan, Y.A, Yuan, Z, Yin, B, Wei, D. | | Deposit date: | 2012-10-09 | | Release date: | 2013-07-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for cofactor and substrate selection by cyanobacterium succinic semialdehyde dehydrogenase
J.Struct.Biol., 182, 2013
|
|
3EUK
 
 | | Crystal structure of MukE-MukF(residues 292-443)-MukB(head domain)-ATPgammaS complex, asymmetric dimer | | Descriptor: | Chromosome partition protein mukB, Linker, Chromosome partition protein mukE, ... | | Authors: | Woo, J.S, Lim, J.H, Shin, H.C, Oh, B.H. | | Deposit date: | 2008-10-10 | | Release date: | 2009-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions.
Cell(Cambridge,Mass.), 136, 2009
|
|
3O6J
 
 | | Crystal Structure of 4-Chlorocatechol Dioxygenase from Rhodococcus opacus 1CP in complex with hydroxyquinol | | Descriptor: | (2R)-3-(PHOSPHONOOXY)-2-(TETRADECANOYLOXY)PROPYL PALMITATE, CHLORIDE ION, Chlorocatechol 1,2-dioxygenase, ... | | Authors: | Ferraroni, M, Briganti, F, Kolomitseva, M, Golovleva, L. | | Deposit date: | 2010-07-29 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray structures of 4-chlorocatechol 1,2-dioxygenase adducts with substituted catechols: new perspectives in the molecular basis of intradiol ring cleaving dioxygenases specificity.
J. Struct. Biol., 181, 2013
|
|
1B8P
 
 | | MALATE DEHYDROGENASE FROM AQUASPIRILLUM ARCTICUM | | Descriptor: | PROTEIN (MALATE DEHYDROGENASE) | | Authors: | Kim, S.Y, Hwang, K.Y, Kim, S.-H, Han, Y.S, Cho, Y. | | Deposit date: | 1999-02-02 | | Release date: | 1999-07-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for cold adaptation. Sequence, biochemical properties, and crystal structure of malate dehydrogenase from a psychrophile Aquaspirillium arcticum.
J.Biol.Chem., 274, 1999
|
|
