4YWN
 
 | |
4NPR
 
 | | Crystal Structure of the Family 12 Xyloglucanase from Aspergillus niveus | | Descriptor: | SULFATE ION, Xyloglucan-specific endo-beta-1,4-glucanase GH12 | | Authors: | Cordeiro, R.L, Santos, C.R, Furtado, G.P, Damasio, A.R.L, Polizeli, M.L.T.M, Ward, R.J, Murakami, M.T. | | Deposit date: | 2013-11-22 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Family 12 Xyloglucanase from Aspergillus niveus
To be Published
|
|
4YUU
 
 | | Crystal structure of oxygen-evolving photosystem II from a red alga | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ago, H, Shen, J.-R. | | Deposit date: | 2015-03-19 | | Release date: | 2016-01-20 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (2.7700038 Å) | | Cite: | Novel Features of Eukaryotic Photosystem II Revealed by Its Crystal Structure Analysis from a Red Alga
J.Biol.Chem., 291, 2016
|
|
4NC5
 
 | | Human sialidase 2 in complex with 2,3-difluorosialic acid (covalent intermediate) | | Descriptor: | 5-acetamido-3,5-dideoxy-3-fluoro-D-erythro-alpha-L-manno-non-2-ulopyranosonic acid, PHOSPHATE ION, Sialidase-2 | | Authors: | Buchini, S, Gallat, F.-X, Greig, I.R, Kim, J.-H, Wakatsuki, S, Chavas, L.M.G, Withers, S.G. | | Deposit date: | 2013-10-24 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.513 Å) | | Cite: | Tuning mechanism-based inactivators of neuraminidases: mechanistic and structural insights.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3ZNU
 
 | | Crystal structure of ClcF in crystal form 2 | | Descriptor: | 1,2-ETHANEDIOL, 5-CHLOROMUCONOLACTONE DEHALOGENASE, CHLORIDE ION, ... | | Authors: | Roth, C, Groening, J.A.D, Kaschabek, S.R, Schloemann, M, Straeter, N. | | Deposit date: | 2013-02-18 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure and Catalytic Mechanism of Chloromuconolactone Dehalogenase Clcf from Rhodococcus Opacus 1Cp.
Mol.Microbiol., 88, 2013
|
|
4NY2
 
 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with acetate ion (ACT) in P 21 | | Descriptor: | ACETATE ION, CALCIUM ION, Deacetylase DA1, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2013-12-10 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.879 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NDI
 
 | | Human Aprataxin (Aptx) AOA1 variant K197Q bound to RNA-DNA, AMP, and Zn - product complex | | Descriptor: | 5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3', 5'-R(P*G)-D(P*TP*TP*AP*TP*GP*AP*TP*TP*C)-3', ADENOSINE MONOPHOSPHATE, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2013-10-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
4ZFC
 
 | | Crystal structure of AKR1C3 complexed with glicazide | | Descriptor: | Aldo-keto reductase family 1 member C3, N-[(3aR,6aS)-hexahydrocyclopenta[c]pyrrol-2(1H)-ylcarbamoyl]-4-methylbenzenesulfonamide, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, Y, Zhrng, X, Hu, X. | | Deposit date: | 2015-04-21 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
4ZMG
 
 | | Crystal structure of Human Dihydroorotate Dehydrogenase (DHODH) with DH03A338 | | Descriptor: | 1-(3,5-difluoro-3'-methoxybiphenyl-4-yl)-3-(1,3-thiazol-5-yl)urea, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Ren, X.L, Zhu, J.S, Zhu, L.L, Li, H.L. | | Deposit date: | 2015-05-04 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Human Dihydroorotate Dehydrogenase (DHODH) with DH03A338
To Be Published
|
|
4ZO9
 
 | | Crystal Structure of mutant (D270A) beta-glucosidase from Listeria innocua in complex with laminaribiose | | Descriptor: | GLYCEROL, Lin1840 protein, MAGNESIUM ION, ... | | Authors: | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | Deposit date: | 2015-05-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
4NGJ
 
 | | Dialyzed HEW lysozyme batch crystallized in 1.0 M RbCl and collected at 100 K | | Descriptor: | CHLORIDE ION, Lysozyme C, RUBIDIUM ION | | Authors: | Benas, P, Legrand, L, Ries-Kautt, M. | | Deposit date: | 2013-11-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Weak protein-cationic co-ion interactions addressed by X-ray crystallography and mass spectrometry.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NMM
 
 | | Crystal Structure of a G12C Oncogenic Variant of Human KRas Bound to a Novel GDP Competitive Covalent Inhibitor | | Descriptor: | 5'-O-[(S)-{[(S)-[2-(acetylamino)ethoxy](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]guanosine, GTPase KRas, MAGNESIUM ION | | Authors: | Hunter, J.C, Gurbani, D, Lim, S.M, Westover, K.D. | | Deposit date: | 2013-11-15 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | In situ selectivity profiling and crystal structure of SML-8-73-1, an active site inhibitor of oncogenic K-Ras G12C.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4YXF
 
 | |
3ZG1
 
 | | NI-BOUND FORM OF M123A MUTANT OF CUPRIAVIDUS METALLIDURANS CH34 CNRXS | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Girard, E, Coves, J. | | Deposit date: | 2012-12-13 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Metal Sensing and Signal Transduction by Cnrx from Cupriavidus Metallidurans Ch34: Role of the Only Methionine Assessed by a Functional, Spectroscopic, and Theoretical Study
Metallomics, 6, 2014
|
|
4NJM
 
 | |
4Z3M
 
 | | X-ray structure of the adduct formed in the reaction between lysozyme and a platinum(II) Complex with S,O Bidentate Ligands (9b) | | Descriptor: | 1,2-ETHANEDIOL, 3-[2-chloranyl-2-[dimethyl(oxidanyl)-{4}-sulfanyl]-4-ethylsulfanyl-1-oxa-3{3}-thia-2{4}-platinacyclohexa-3,5-dien-6-yl]phenol, DIMETHYL SULFOXIDE, ... | | Authors: | Merlino, A. | | Deposit date: | 2015-03-31 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Platinum(II) Complexes with O,S Bidentate Ligands: Biophysical Characterization, Antiproliferative Activity, and Crystallographic Evidence of Protein Binding.
Inorg.Chem., 54, 2015
|
|
4NWU
 
 | |
4Z40
 
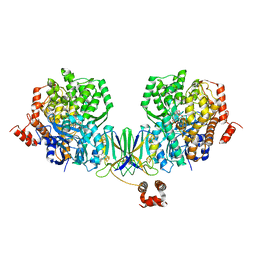 | | Active site complex BamBC of Benzoyl Coenzyme A reductase as isolated | | Descriptor: | Benzoyl-CoA reductase, putative, IRON/SULFUR CLUSTER, ... | | Authors: | Weinert, T, Kung, J.W, Weidenweber, S, Huwiler, S.G, Boll, M, Ermler, U. | | Deposit date: | 2015-04-01 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of enzymatic benzene ring reduction.
Nat.Chem.Biol., 11, 2015
|
|
4B3Q
 
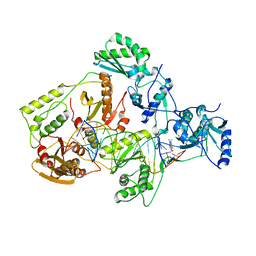 | | Structures of HIV-1 RT and RNA-DNA Complex Reveal a Unique RT Conformation and Substrate Interface | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, P51 RT, PRIMER DNA, ... | | Authors: | Lapkouski, M, Tian, L, Miller, J.T, Le Grice, S.F.J, Yang, W. | | Deposit date: | 2012-07-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Complexes of HIV-1 RT, Nnrti and RNA/DNA Hybrid Reveal a Structure Compatible with RNA Degradation
Nat.Struct.Mol.Biol., 20, 2013
|
|
4NWT
 
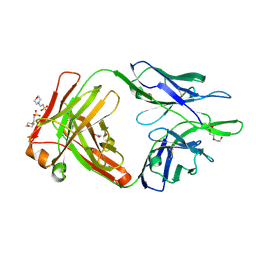 | | Crystal structure of the anti-human NGF Fab APE1531 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, APE1531 Ab Fab heavy chain, ... | | Authors: | Verdino, P, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2013-12-06 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Nucleotide insertions and deletions complement point mutations to massively expand the diversity created by somatic hypermutation of antibodies.
J.Biol.Chem., 289, 2014
|
|
4ZOE
 
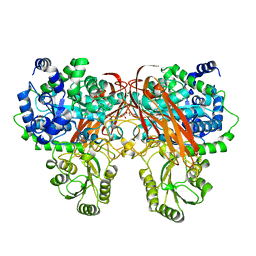 | | Crystal Structure of beta-glucosidase from Listeria innocua | | Descriptor: | GLYCEROL, Lin1840 protein, MAGNESIUM ION | | Authors: | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | Deposit date: | 2015-05-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
4Z82
 
 | | Cysteine bound rat cysteine dioxygenase C164S variant at pH 8.1 | | Descriptor: | CYSTEINE, Cysteine dioxygenase type 1, FE (II) ION | | Authors: | Fellner, M, Tchesnokov, E.P, Siakkou, E, Rutledge, M.T, Kanitz, M, Jameson, G.N.L, Wilbanks, S.M. | | Deposit date: | 2015-04-08 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Influence of cysteine 164 on active site structure in rat cysteine dioxygenase.
J.Biol.Inorg.Chem., 21, 2016
|
|
4Z85
 
 | |
4NYU
 
 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with acetate ion (ACT) in C 2 2 21 | | Descriptor: | ACETATE ION, CALCIUM ION, Deacetylase DA1, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2013-12-11 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4Z9G
 
 | | Crystal structure of human corticotropin-releasing factor receptor 1 (CRF1R) in complex with the antagonist CP-376395 in a hexagonal setting with translational non-crystallographic symmetry | | Descriptor: | 3,6-dimethyl-N-(pentan-3-yl)-2-(2,4,6-trimethylphenoxy)pyridin-4-amine, Corticotropin-releasing factor receptor 1,Lysozyme,Corticotropin-releasing factor receptor 1, OLEIC ACID, ... | | Authors: | Dore, A.S, Bortolato, A, Hollenstein, K, Cheng, R.K.Y, Read, R.J, Marshall, F.H. | | Deposit date: | 2015-04-10 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.183 Å) | | Cite: | Decoding Corticotropin-Releasing Factor Receptor Type 1 Crystal Structures.
Curr Mol Pharmacol, 10, 2017
|
|
