4WVB
 
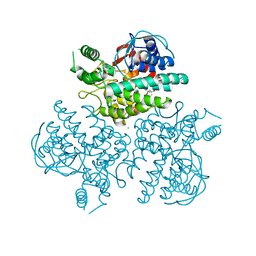 | | Crystal structure of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8 in complex with glucose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Uncharacterized protein, ... | | Authors: | Miyazaki, T, Ichikawa, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2014-11-05 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure and substrate-binding mode of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8.
J.Struct.Biol., 190, 2015
|
|
4AND
 
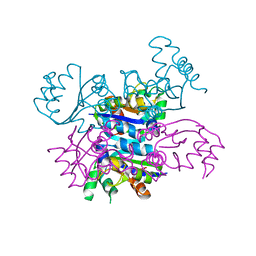 | | CRYSTAL FORM II OF THE D93N MUTANT OF NUCLEOSIDE DIPHOSPHATE KINASE FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Georgescauld, F, Moynie, L, Habersetzer, J, Lascu, I, Dautant, A. | | Deposit date: | 2012-03-16 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.808 Å) | | Cite: | Intersubunit Ionic Interactions Stabilize the Nucleoside Diphosphate Kinase of Mycobacterium Tuberculosis.
Plos One, 8, 2013
|
|
4X4P
 
 | |
4NEB
 
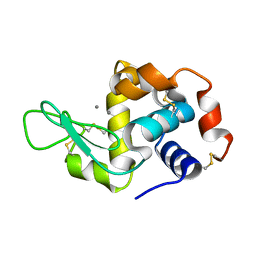 | | Previously de-ionized HEW lysozyme batch crystallized in 0.5 M MnCl2 | | Descriptor: | CHLORIDE ION, Lysozyme C, MANGANESE (II) ION | | Authors: | Benas, P, Legrand, L, Ries-Kautt, M. | | Deposit date: | 2013-10-29 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Weak protein-cationic co-ion interactions addressed by X-ray crystallography and mass spectrometry.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NM9
 
 | |
4ND5
 
 | |
4NHL
 
 | | Crystal structure of Tpa1p from Saccharomyces cerevisiae, termination and polyadenylation protein 1, in complex with N-oxalylglycine (NOG) | | Descriptor: | MANGANESE (II) ION, N-OXALYLGLYCINE, PKHD-type hydroxylase TPA1 | | Authors: | Scotti, J.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2013-11-05 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structure of the Ribosomal Oxygenase OGFOD1 Provides Insights into the Regio- and Stereoselectivity of Prolyl Hydroxylases.
Structure, 23, 2015
|
|
4NFY
 
 | |
4NZ1
 
 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with DI(N-ACETYL-D-GLUCOSAMINE) (CBS) in P 21 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2013-12-11 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4X28
 
 | | Crystal structure of the ChsE4-ChsE5 complex from Mycobacterium tuberculosis | | Descriptor: | Acyl-CoA dehydrogenase, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE | | Authors: | Guja, K.E, Yang, M, Sampson, N, Garcia-Diaz, M. | | Deposit date: | 2014-11-26 | | Release date: | 2015-02-18 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Unraveling Cholesterol Catabolism in Mycobacterium tuberculosis: ChsE4-ChsE5 alpha 2 beta 2 Acyl-CoA Dehydrogenase Initiates beta-Oxidation of 3-Oxo-cholest-4-en-26-oyl CoA.
Acs Infect Dis., 1, 2015
|
|
4AFB
 
 | | Crystal Structure of subtype-switched Epithelial Adhesin 1 to 3 A domain (Epa1to3A) from Candida glabrata in complex with glycerol | | Descriptor: | CALCIUM ION, EPA1P, GLYCEROL | | Authors: | Maestre-Reyna, M, Diderrich, R, Veelders, M.S, Eulenburg, G, Kalugin, V, Brueckner, S, Keller, P, Rupp, S, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2012-01-18 | | Release date: | 2012-10-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Promiscuity and Specificity During Candida Glabrata Invasion of Host Epithelia.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4O59
 
 | | Co-enzyme Induced Conformational Changes in Bovine Eye Glyceraldehyde 3-Phosphate Dehydrogenase | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Baker, B.Y, Shi, W, Wang, B, Palczewski, K. | | Deposit date: | 2013-12-19 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | High-resolution crystal structures of the photoreceptor glyceraldehyde 3-phosphate dehydrogenase (GAPDH) with three and four-bound NAD molecules.
Protein Sci., 23, 2014
|
|
4A1F
 
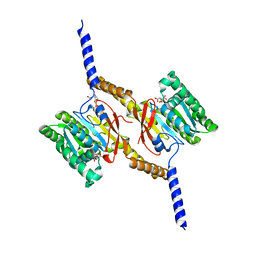 | | Crystal structure of C-terminal domain of Helicobacter pylori DnaB Helicase | | Descriptor: | CITRATE ANION, REPLICATIVE DNA HELICASE | | Authors: | Stelter, M, Kapp, U, Timmins, J, Terradot, L. | | Deposit date: | 2011-09-14 | | Release date: | 2012-03-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Architecture of a Dodecameric Bacterial Replicative Helicase.
Structure, 20, 2012
|
|
4ND4
 
 | |
4ACC
 
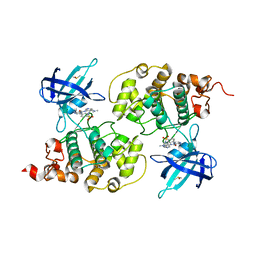 | | GSK3b in complex with inhibitor | | Descriptor: | 3-AMINO-6-(4-{[2-(DIMETHYLAMINO)ETHYL]SULFAMOYL}PHENYL)-N-PYRIDIN-3-YLPYRAZINE-2-CARBOXAMIDE, DIMETHYL SULFOXIDE, GLYCOGEN SYNTHASE KINASE-3 BETA | | Authors: | Xue, Y, Ormo, M. | | Deposit date: | 2011-12-14 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery of novel potent and highly selective glycogen synthase kinase-3 beta (GSK3 beta ) inhibitors for Alzheimer's disease: design, synthesis, and characterization of pyrazines.
J. Med. Chem., 55, 2012
|
|
4WNC
 
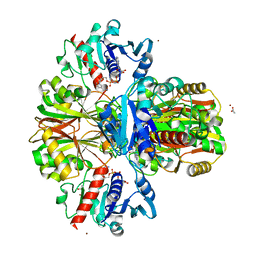 | | Crystal structure of human wild-type GAPDH at 1.99 angstroms resolution | | Descriptor: | ACETATE ION, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Garcin, E.D. | | Deposit date: | 2014-10-11 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A Dimer Interface Mutation in Glyceraldehyde-3-Phosphate Dehydrogenase Regulates Its Binding to AU-rich RNA.
J.Biol.Chem., 290, 2015
|
|
4WPN
 
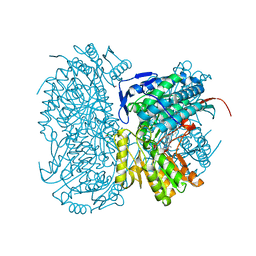 | | Structure of human ALDH1A1 with inhibitor CM053 | | Descriptor: | 1-{[1,3-dimethyl-7-(3-methylbutyl)-2,6-dioxo-2,3,6,7-tetrahydro-1H-purin-8-yl]methyl}piperidine-4-carboxamide, CHLORIDE ION, Retinal dehydrogenase 1, ... | | Authors: | Morgan, C.A, Hurley, T.D. | | Deposit date: | 2014-10-20 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Characterization of Two Distinct Structural Classes of Selective Aldehyde Dehydrogenase 1A1 Inhibitors.
J.Med.Chem., 58, 2015
|
|
7POZ
 
 | |
4NS3
 
 | |
4ZOB
 
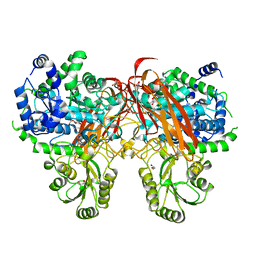 | | Crystal Structure of beta-glucosidase from Listeria innocua in complex with gluconolactone | | Descriptor: | D-glucono-1,5-lactone, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | Deposit date: | 2015-05-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
4NPI
 
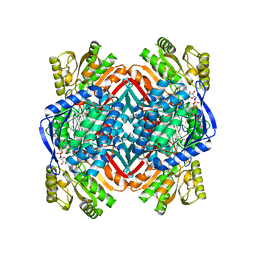 | | 1.94 Angstroms X-ray crystal structure of NAD- and intermediate- bound alpha-aminomuconate-epsilon-semialdehyde dehydrogenase from Pseudomonas fluorescens | | Descriptor: | (2Z,4E)-2-hydroxy-6-oxohexa-2,4-dienoic acid, 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Huo, L, Davis, I, Liu, F, Iwaki, H, Hasegawa, Y, Liu, A. | | Deposit date: | 2013-11-21 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action.
Nat Commun, 6, 2015
|
|
4YYE
 
 | |
4YZP
 
 | |
4NU6
 
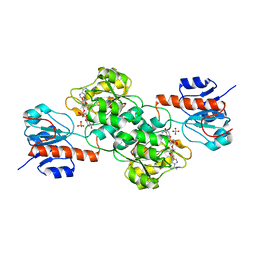 | | Crystal Structure of PTDH R301K | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Phosphonate dehydrogenase, SULFATE ION | | Authors: | Nair, S.K, Chekan, J.R. | | Deposit date: | 2013-12-03 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Chemical rescue and inhibition studies to determine the role of arg301 in phosphite dehydrogenase.
Plos One, 9, 2014
|
|
4Z24
 
 | | Mimivirus R135 (residues 51-702) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GMC-type oxidoreductase R135 | | Authors: | Klose, T, Rossmann, M.G. | | Deposit date: | 2015-03-28 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mimivirus Enzyme that Participates in Viral Entry.
Structure, 23, 2015
|
|
