1GNQ
 
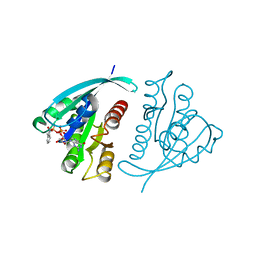 | | X-RAY CRYSTAL STRUCTURE ANALYSIS OF THE CATALYTIC DOMAIN OF THE ONCOGENE PRODUCT P21H-RAS COMPLEXED WITH CAGED GTP AND MANT DGPPNHP | | Descriptor: | C-H-RAS P21 PROTEIN, GUANOSINE 5'-TRIPHOSPHATE P3-[1-(2-NITROPHENYL)ETHYL ESTER], MAGNESIUM ION | | Authors: | Scheidig, A, Franken, S.M, Corrie, J.E.T, Reid, G.P, Wittinghofer, A, Pai, E.F, Goody, R.S. | | Deposit date: | 1995-05-11 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystal structure analysis of the catalytic domain of the oncogene product p21H-ras complexed with caged GTP and mant dGppNHp.
J.Mol.Biol., 253, 1995
|
|
1AUY
 
 | |
1GRQ
 
 | |
1H04
 
 | | Human CD55 domains 3 & 4 | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR, NICKEL (II) ION | | Authors: | Williams, P, Chaudhry, Y, Goodfellow, I, Billington, J, Spiller, B, Evans, D.J, Lea, S.M. | | Deposit date: | 2002-06-11 | | Release date: | 2003-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping Cd55 Function. The Structure of Two Pathogen-Binding Domains at 1.7 A
J.Biol.Chem., 278, 2003
|
|
1H2P
 
 | | Human CD55 domains 3 & 4 | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR | | Authors: | Williams, P, Chaudhry, Y, Goodfellow, I, Billington, J, Spiller, B, Evans, D.J, Lea, S.M. | | Deposit date: | 2002-08-13 | | Release date: | 2003-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mapping Cd55 Function. The Structure of Two Pathogen-Binding Domains at 1.7 A
J.Biol.Chem., 278, 2003
|
|
1H03
 
 | | Human CD55 domains 3 & 4 | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR | | Authors: | Williams, P, Chaudhry, Y, Goodfellow, I, Billington, J, Spiller, B, Evans, D.J, Lea, S.M. | | Deposit date: | 2002-06-11 | | Release date: | 2003-03-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mapping Cd55 Function. The Structure of Two Pathogen-Binding Domains at 1.7 A
J.Biol.Chem., 278, 2003
|
|
8Q6K
 
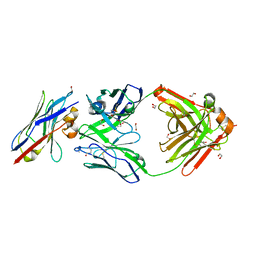 | | Human IgD Fab in complex with an orthosteric inhibitor of Phl p 7 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Human IgD Fab heavy chain, ... | | Authors: | Davies, A.M, Vester, S.K, McDonnell, J.M. | | Deposit date: | 2023-08-11 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Expanding the Anti-Phl p 7 Antibody Toolkit: An Anti-Idiotype Nanobody Inhibitor.
Antibodies, 12, 2023
|
|
8R1F
 
 | | Monomeric E6AP-E6-p53 ternary complex | | Descriptor: | Cellular tumor antigen p53, Ubiquitin-like protein SMT3,Protein E6, Ubiquitin-protein ligase E3A, ... | | Authors: | Sandate, C.R, Chakraborty, D, Kater, L, Kempf, G, Thoma, N.H. | | Deposit date: | 2023-11-01 | | Release date: | 2023-12-06 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural insights into viral hijacking of p53 by E6 and E6AP
Biorxiv, 2023
|
|
1ARV
 
 | | CRYSTAL STRUCTURES OF CYANIDE-AND TRIIODIDE-BOUND FORMS OF ARTHROMYCES RAMOSUS PEROXIDASE AT DIFFERENT PH VALUES. PERTURBATIONS OF ACTIVE SITE RESIDUES AND THEIR IMPLICATION IN ENZYME CATALYSIS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYANIDE ION, ... | | Authors: | Fukuyama, K, Kunishima, N, Amada, F. | | Deposit date: | 1995-04-25 | | Release date: | 1996-01-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of cyanide- and triiodide-bound forms of Arthromyces ramosus peroxidase at different pH values. Perturbations of active site residues and their implication in enzyme catalysis.
J.Biol.Chem., 270, 1995
|
|
1GP4
 
 | | Anthocyanidin synthase from Arabidopsis thaliana (selenomethionine substituted) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-OXOGLUTARIC ACID, ANTHOCYANIDIN SYNTHASE | | Authors: | Wilmouth, R.C, Turnbull, J.J, Welford, R.W.D, Clifton, I.J, Prescott, A.G, Schofield, C.J. | | Deposit date: | 2001-10-30 | | Release date: | 2002-02-21 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Mechanism of Anthocyanidin Synthase from Arabidopsis Thaliana.
Structure, 10, 2002
|
|
1ATJ
 
 | | RECOMBINANT HORSERADISH PEROXIDASE C1A | | Descriptor: | CALCIUM ION, PEROXIDASE C1A, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gajhede, M, Schuller, D.J, Henriksen, A, Smith, A.T, Poulos, T.L. | | Deposit date: | 1997-08-14 | | Release date: | 1998-02-04 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of horseradish peroxidase C at 2.15 A resolution.
Nat.Struct.Biol., 4, 1997
|
|
1HBP
 
 | |
5K4M
 
 | |
1ARU
 
 | | CRYSTAL STRUCTURES OF CYANIDE-AND TRIIODIDE-BOUND FORMS OF ARTHROMYCES RAMOSUS PEROXIDASE AT DIFFERENT PH VALUES. PERTURBATIONS OF ACTIVE SITE RESIDUES AND THEIR IMPLICATION IN ENZYME CATALYSIS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYANIDE ION, ... | | Authors: | Fukuyama, K, Kunishima, N, Amada, F. | | Deposit date: | 1995-04-25 | | Release date: | 1996-01-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of cyanide- and triiodide-bound forms of Arthromyces ramosus peroxidase at different pH values. Perturbations of active site residues and their implication in enzyme catalysis.
J.Biol.Chem., 270, 1995
|
|
1FDP
 
 | | PROENZYME OF HUMAN COMPLEMENT FACTOR D, RECOMBINANT PROFACTOR D | | Descriptor: | PROENZYME OF COMPLEMENT FACTOR D | | Authors: | Jing, H, Macon, K.J, Moore, D, Delucas, L.J, Volanakis, J.E, Narayana, S.V.L. | | Deposit date: | 1998-12-03 | | Release date: | 1999-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of profactor D activation: from a highly flexible zymogen to a novel self-inhibited serine protease, complement factor D.
Embo J., 18, 1999
|
|
1CLY
 
 | |
1CJ4
 
 | | MUTANT Q34T OF PARA-HYDROXYBENZOATE HYDROXYLASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID, PROTEIN (P-HYDROXYBENZOATE HYDROXYLASE) | | Authors: | Eppink, M.H.M, Overkamp, K.M, Schreuder, H.A, Van Berkel, W.J.H. | | Deposit date: | 1999-04-21 | | Release date: | 1999-04-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Switch of coenzyme specificity of p-hydroxybenzoate hydroxylase.
J.Mol.Biol., 292, 1999
|
|
1XPS
 
 | |
3TOS
 
 | | Crystal Structure of CalS11, Calicheamicin Methyltransferase | | Descriptor: | 1,2-ETHANEDIOL, CalS11, GLUTAMIC ACID, ... | | Authors: | Chang, A, Aceti, D.J, Beebe, E.T, Makino, S.-I, Wrobel, R.L, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2011-09-06 | | Release date: | 2011-10-05 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of CalS11, Calicheamicin methyltransferase
To be Published
|
|
1CRZ
 
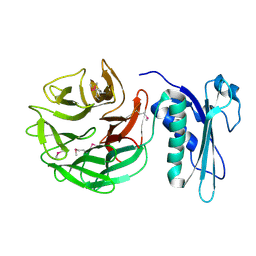 | | CRYSTAL STRUCTURE OF THE E. COLI TOLB PROTEIN | | Descriptor: | TOLB PROTEIN | | Authors: | Abergel, C, Bouveret, E, Claverie, J.-M, Brown, K, Rigal, A, Lazdunski, C, Benedetti, H. | | Deposit date: | 1999-08-16 | | Release date: | 2000-08-16 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the Escherichia coli TolB protein determined by MAD methods at 1.95 A resolution.
Structure Fold.Des., 7, 1999
|
|
1CYW
 
 | | QUINOL OXIDASE (PERIPLASMIC FRAGMENT OF SUBUNIT II) (CYOA) | | Descriptor: | CYOA | | Authors: | Wilmanns, M, Lappalainen, P, Kelly, M, Sauer-Eriksson, E, Saraste, M. | | Deposit date: | 1995-08-22 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the membrane-exposed domain from a respiratory quinol oxidase complex with an engineered dinuclear copper center.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
5JQJ
 
 | |
1RJC
 
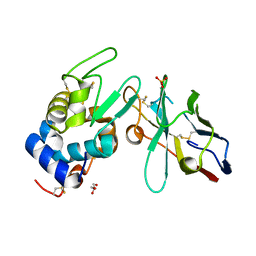 | | Crystal structure of the camelid single domain antibody cAb-Lys2 in complex with hen egg white lysozyme | | Descriptor: | GLYCEROL, Lysozyme C, PHOSPHATE ION, ... | | Authors: | De Genst, E, Silence, K, Ghahroudi, M.A, Decanniere, K, Loris, R, Kinne, J, Wyns, L, Muyldermans, S. | | Deposit date: | 2003-11-19 | | Release date: | 2005-02-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Strong in vivo maturation compensates for structurally restricted H3 loops in antibody repertoires.
J.Biol.Chem., 280, 2005
|
|
1I3W
 
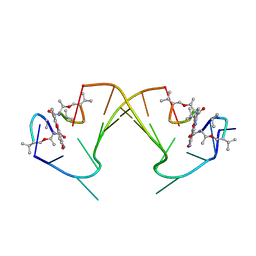 | | ACTINOMYCIN D BINDING TO CGATCGATCG | | Descriptor: | 5'-D(*C*GP*AP*TP*CP*GP*AP*(BRU)P*CP*GP)-3', ACTINOMYCIN D | | Authors: | Robinson, H, Gao, Y.-G, Yang, X.-L, Sanishvili, R, Joachimiak, A, Wang, A.H.-J. | | Deposit date: | 2001-02-17 | | Release date: | 2001-05-21 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic Analysis of a Novel Complex of Actinomycin D Bound to the DNA Decamer Cgatcgatcg.
Biochemistry, 40, 2001
|
|
1I4P
 
 | |
