8V2W
 
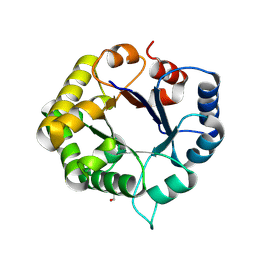 | | Crystal Structure of the ancestral triosephosphate isomerase reconstruction of the last opisthokont common ancestor obtained by Bayesian inference | | Descriptor: | GLYCEROL, Triosephosphate isomerase | | Authors: | Perez-Nino, J.A, Rodriguez-Romero, A, Guerra, Y, Fernandez-Velasco, D.A. | | Deposit date: | 2023-11-24 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Stable monomers in the ancestral sequence reconstruction of the last opisthokont common ancestor of dimeric triosephosphate isomerase.
Protein Sci., 33, 2024
|
|
8DND
 
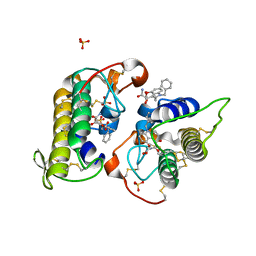 | |
8F3A
 
 | | HIV-1 gp41 coiled-coil pocket IQN17 | | Descriptor: | ACETIC ACID, CHLORIDE ION, IQN17 | | Authors: | Bruun, T.U.J, Tang, S, Fernandez, D, Kim, P.S. | | Deposit date: | 2022-11-09 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-guided stabilization improves the ability of the HIV-1 gp41 hydrophobic pocket to elicit neutralizing antibodies.
J.Biol.Chem., 299, 2023
|
|
8F3B
 
 | |
8DZK
 
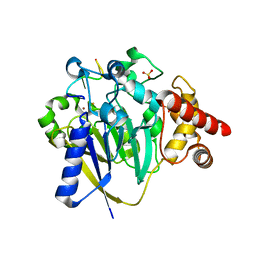 | | Dbr1 in complex with 5-mer cleavage product | | Descriptor: | FE (II) ION, RNA (5'-R(P*(G46)P*UP*GP*UP*U)-3'), RNA lariat debranching enzyme, ... | | Authors: | Clark, N.E, Taylor, A.B. | | Deposit date: | 2022-08-08 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the RNA Lariat Debranching Enzyme Dbr1 with Hydrolyzed Phosphorothioate RNA Product.
Biochemistry, 61, 2022
|
|
8VKD
 
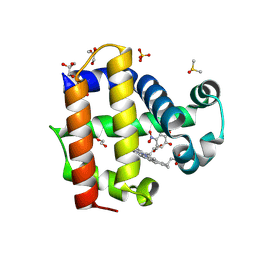 | | Crystal structure of dehaloperoxidase A in complex with substrate 4-nitrocatechol | | Descriptor: | 4-NITROCATECHOL, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Ghiladi, R.A, Franzen, S. | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Comparison of Substrate Binding Sites in Dehaloperoxidase A and B.
Biochemistry, 63, 2024
|
|
6ZVF
 
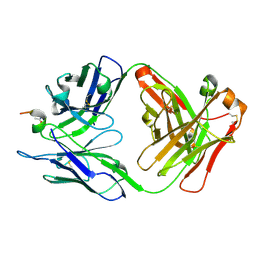 | |
8VKC
 
 | | Crystal structure of dehaloperoxidase A in complex with substrate 4-nitrophenol | | Descriptor: | Dehaloperoxidase A, GLYCEROL, P-NITROPHENOL, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Ghiladi, R.A, Franzen, S. | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Comparison of Substrate Binding Sites in Dehaloperoxidase A and B.
Biochemistry, 63, 2024
|
|
8V2X
 
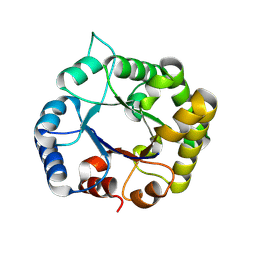 | |
7A70
 
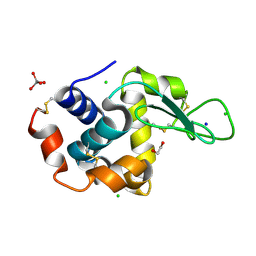 | | HEW lysozyme in complex with Ti(OH)4 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme, ... | | Authors: | Calderone, V, Gigli, L, Ravera, E, Luchinat, C. | | Deposit date: | 2020-08-27 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | On the Mechanism of Bioinspired Formation of Inorganic Oxides: Structural Evidence of the Electrostatic Nature of the Interaction between a Mononuclear Inorganic Precursor and Lysozyme.
Biomolecules, 11, 2020
|
|
8V09
 
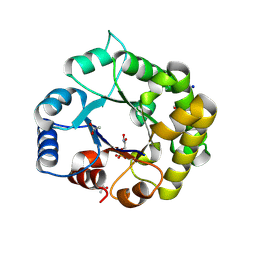 | | Crystal Structure of the reconstruction of the ancestral triosephosphate isomerase of the last opisthokont common ancestor obtained by bayesian inference with PGH | | Descriptor: | ACETIC ACID, FORMIC ACID, PHOSPHOGLYCOLOHYDROXAMIC ACID, ... | | Authors: | Perez-Nino, J.A, Rodriguez-Romero, A, Guerra-Borrego, Y, Fernandez-Velasco, D.A. | | Deposit date: | 2023-11-17 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Stable monomers in the ancestral sequence reconstruction of the last opisthokont common ancestor of dimeric triosephosphate isomerase.
Protein Sci., 33, 2024
|
|
8F5K
 
 | |
8F5L
 
 | | Azurin from Pseudomonas aeruginosa, Y72F/Y108F/F110L mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Azurin, COPPER (II) ION | | Authors: | Zeug, M, Offenbacher, A.R, Choe, J. | | Deposit date: | 2022-11-14 | | Release date: | 2023-01-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Electrochemical and Structural Study of the Buried Tryptophan in Azurin: Effects of Hydration and Polarity on the Redox Potential of W48.
J.Phys.Chem.B, 127, 2023
|
|
8VSK
 
 | | Crystal structure of Dehaloperoxidase A in complex with substrate 2,4-dibromophenol | | Descriptor: | 2,4-bis(bromanyl)phenol, DI(HYDROXYETHYL)ETHER, Dehaloperoxidase A, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Ghiladi, R.A, Franzen, S. | | Deposit date: | 2024-01-24 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.515 Å) | | Cite: | Structural Comparison of Substrate Binding Sites in Dehaloperoxidase A and B.
Biochemistry, 63, 2024
|
|
8WZU
 
 | |
8EWV
 
 | | DNA-encoded library (DEL)-enabled discovery of proximity inducing small molecules | | Descriptor: | Bromodomain-containing protein 4, Elongin-B, Elongin-C, ... | | Authors: | Schreiber, S.L, Shu, W, Ma, X, Michaud, G, Bonazzi, S, Berst, F. | | Deposit date: | 2022-10-24 | | Release date: | 2023-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | DNA-encoded library-enabled discovery of proximity-inducing small molecules.
Nat.Chem.Biol., 20, 2024
|
|
8ETQ
 
 | |
8TWR
 
 | | Influenza A virus (A/Aichi/2/1968(H3N2) nucleoprotein mutant - 2-7 deleted, P283S, R416A | | Descriptor: | Nucleoprotein, SODIUM ION | | Authors: | Yoon, J, Zhang, Y.M, Grant, R.A, Shoulders, M.D. | | Deposit date: | 2023-08-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | The immune-evasive proline-283 substitution in influenza nucleoprotein increases aggregation propensity without altering the native structure.
Sci Adv, 10, 2024
|
|
8VYL
 
 | |
8VZR
 
 | | Crystal structure of dehaloperoxidase A in complex with substrate 4-bromo-o-cresol | | Descriptor: | 4-bromo-2-methylphenol, DI(HYDROXYETHYL)ETHER, Dehaloperoxidase A, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Ghiladi, R.A, Franzen, S. | | Deposit date: | 2024-02-12 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Comparison of Substrate Binding Sites in Dehaloperoxidase A and B.
Biochemistry, 63, 2024
|
|
8W08
 
 | | Crystal Structure of the worst case reconstruction of the ancestral triosephosphate isomerase of the last opisthokont common ancestor obtained by maximum likelihood | | Descriptor: | FLUORIDE ION, Triosephosphate isomerase | | Authors: | Perez-Nino, J.A, Rodriguez-Romero, A, Guerra-Borrego, Y, Fernandez-Velasco, D.A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Stable monomers in the ancestral sequence reconstruction of the last opisthokont common ancestor of dimeric triosephosphate isomerase.
Protein Sci., 33, 2024
|
|
8EK4
 
 | |
8UGC
 
 | |
8W05
 
 | |
8W06
 
 | | Crystal Structure of the reconstruction of the ancestral triosephosphate isomerase of the last opisthokont common ancestor obtained by maximum likelihood with PGH | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ACETIC ACID, ... | | Authors: | Perez-Nino, J.A, Rodriguez-Romero, A, Guerra-Borrego, Y, Fernandez-Velasco, D.A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Stable monomers in the ancestral sequence reconstruction of the last opisthokont common ancestor of dimeric triosephosphate isomerase.
Protein Sci., 33, 2024
|
|
