8HTD
 
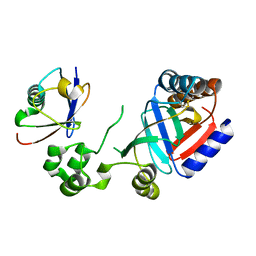 | | Crystal structure of an effector from Chromobacterium violaceum in complex with ubiquitin | | Descriptor: | NAD(+)--protein-threonine ADP-ribosyltransferase, Ubiquitin | | Authors: | Tan, J, Wang, X, Zhou, Y, Zhu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-22 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Molecular basis of threonine ADP-ribosylation of ubiquitin by bacterial ARTs.
Nat.Chem.Biol., 20, 2024
|
|
8I01
 
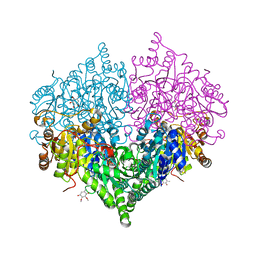 | | Crystal structure of Escherichia coli glyoxylate carboligase | | Descriptor: | 2,3-DIMETHOXY-5-METHYL-1,4-BENZOQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, Glyoxylate carboligase, ... | | Authors: | Kim, J.H, Kim, J.S. | | Deposit date: | 2023-01-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Engineering of two thiamine diphosphate-dependent enzymes for the regioselective condensation of C1-formaldehyde into C4-erythrulose.
Int.J.Biol.Macromol., 253, 2023
|
|
8I07
 
 | |
8I08
 
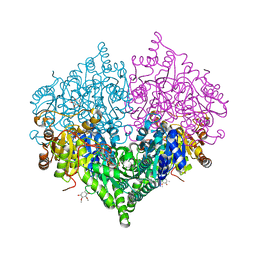 | | Crystal structure of Escherichia coli glyoxylate carboligase quadruple mutant | | Descriptor: | 2,3-DIMETHOXY-5-METHYL-1,4-BENZOQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, Glyoxylate carboligase, ... | | Authors: | Kim, J.H, Kim, J.S. | | Deposit date: | 2023-01-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Engineering of two thiamine diphosphate-dependent enzymes for the regioselective condensation of C1-formaldehyde into C4-erythrulose.
Int.J.Biol.Macromol., 253, 2023
|
|
8HTC
 
 | | Crystal structure of a SeMet-labeled effector from Chromobacterium violaceum in complex with Ubiquitin | | Descriptor: | NAD(+)--protein-threonine ADP-ribosyltransferase, Ubiquitin-40S ribosomal protein S27a (Fragment) | | Authors: | Tan, J, Wang, X, Zhou, Y, Zhu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-22 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis of threonine ADP-ribosylation of ubiquitin by bacterial ARTs.
Nat.Chem.Biol., 20, 2024
|
|
8HTE
 
 | | Crystal structure of an effector mutant in complex with ubiquitin | | Descriptor: | GLYCEROL, NAD(+)--protein-threonine ADP-ribosyltransferase, NICOTINAMIDE, ... | | Authors: | Tan, J, Wang, X, Zhou, Y, Zhu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-22 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | Molecular basis of threonine ADP-ribosylation of ubiquitin by bacterial ARTs.
Nat.Chem.Biol., 20, 2024
|
|
8HTF
 
 | | Crystal structure of an effector in complex with ubiquitin | | Descriptor: | NAD(+)--protein-threonine ADP-ribosyltransferase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Ubiquitin-40S ribosomal protein S27a (Fragment) | | Authors: | Tan, J, Wang, X, Zhou, Y, Zhu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-22 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Molecular basis of threonine ADP-ribosylation of ubiquitin by bacterial ARTs.
Nat.Chem.Biol., 20, 2024
|
|
8I05
 
 | | Crystal structure of Escherichia coli glyoxylate carboligase double mutant | | Descriptor: | 2,3-DIMETHOXY-5-METHYL-1,4-BENZOQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, Glyoxylate carboligase, ... | | Authors: | Kim, J.H, Kim, J.S. | | Deposit date: | 2023-01-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Engineering of two thiamine diphosphate-dependent enzymes for the regioselective condensation of C1-formaldehyde into C4-erythrulose.
Int.J.Biol.Macromol., 253, 2023
|
|
8I6U
 
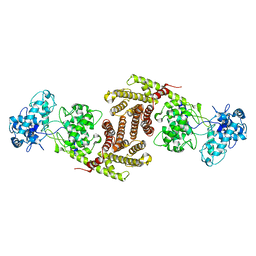 | | The cryo-EM structure of OsCyc1 dimer state | | Descriptor: | Syn-copalyl diphosphate synthase, chloroplastic | | Authors: | Ma, X.L, Xu, H.F, Tong, Y.R, Luo, Y.F, Dong, Q.H, Jiang, T. | | Deposit date: | 2023-01-29 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structural and functional investigations of syn-copalyl diphosphate synthase from Oryza sativa.
Commun Chem, 6, 2023
|
|
8I6T
 
 | | The cryo-EM structure of OsCyc1 hexamer state | | Descriptor: | Syn-copalyl diphosphate synthase, chloroplastic | | Authors: | Ma, X.L, Xu, H.F, Tong, Y.R, Luo, Y.F, Dong, Q.H, Jiang, T. | | Deposit date: | 2023-01-29 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural and functional investigations of syn-copalyl diphosphate synthase from Oryza sativa.
Commun Chem, 6, 2023
|
|
8I6P
 
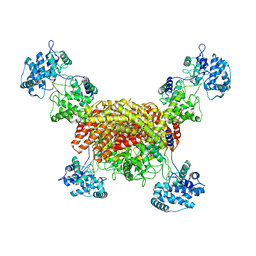 | | The cryo-EM structure of OsCyc1 tetramer state | | Descriptor: | Syn-copalyl diphosphate synthase, chloroplastic | | Authors: | Ma, X.L, Xu, H.F, Tong, Y.R, Luo, Y.F, Dong, Q.H, Jiang, T. | | Deposit date: | 2023-01-29 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural and functional investigations of syn-copalyl diphosphate synthase from Oryza sativa.
Commun Chem, 6, 2023
|
|
8IH5
 
 | |
8HT0
 
 | |
8IB8
 
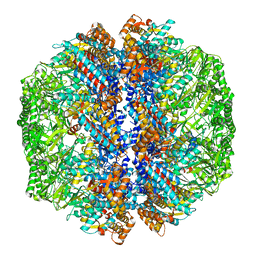 | | Human TRiC-PhLP2A-actin complex in the closed state | | Descriptor: | ACTB protein (Fragment), Phosducin-like protein 3, T-complex protein 1 subunit alpha, ... | | Authors: | Roh, S.H, Park, J, Kim, H, Lim, S. | | Deposit date: | 2023-02-09 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.42 Å) | | Cite: | A structural vista of phosducin-like PhLP2A-chaperonin TRiC cooperation during the ATP-driven folding cycle.
Nat Commun, 15, 2024
|
|
8HUC
 
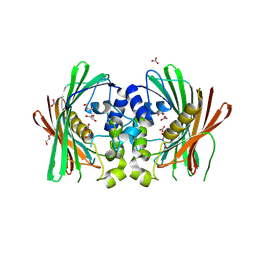 | | Crystal structure of PaIch (Pec1) | | Descriptor: | GLYCEROL, MaoC_dehydrat_N domain-containing protein, NITRATE ION | | Authors: | Pramanik, A, Datta, S. | | Deposit date: | 2022-12-23 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Structural and functional insights of itaconyl-CoA hydratase from Pseudomonas aeruginosa highlight a novel N-terminal hotdog fold.
Febs Lett., 598, 2024
|
|
8HWA
 
 | | D5 ATP-ADP-Apo-ssDNA IS1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Li, Y.N, Zhu, J, Guo, Y.Y, Yan, R.H. | | Deposit date: | 2022-12-29 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insight into the assembly and working mechanism of helicase-primase D5 from Mpox virus.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HWY
 
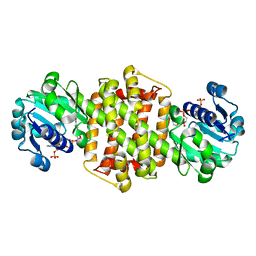 | | Ancestral imine reductase mutant N559_M6 | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ancestral imine reductase mutant N559_M6 | | Authors: | Zhu, X.X. | | Deposit date: | 2023-01-03 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The evolution of stereoselectivity in imine reductase
To Be Published
|
|
8HWK
 
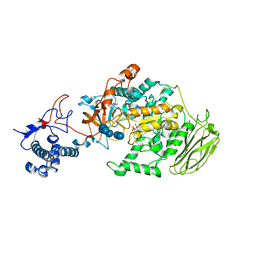 | | Limosilactobacillus reuteri N1 GtfB-maltohexaose | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Dong, J.J, Bai, Y.X. | | Deposit date: | 2022-12-30 | | Release date: | 2024-01-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights into the Structure-Function Relationship of GH70 GtfB alpha-Glucanotransferases from the Crystal Structure and Molecular Dynamic Simulation of a Newly Characterized Limosilactobacillus reuteri N1 GtfB Enzyme.
J.Agric.Food Chem., 72, 2024
|
|
8HZ5
 
 | |
8HWB
 
 | | D5 ATP-ADP-Apo-ssDNA IS2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Li, Y.N, Zhu, J, Guo, Y.Y, Yan, R.H. | | Deposit date: | 2022-12-29 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insight into the assembly and working mechanism of helicase-primase D5 from Mpox virus.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8IJK
 
 | | human KCNQ2-CaM-Ebio1 complex in the presence of PIP2 | | Descriptor: | Calmodulin-1, N-(1,2-dihydroacenaphthylen-5-yl)-4-fluoranyl-benzamide, Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Ma, D, Guo, J. | | Deposit date: | 2023-02-27 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A small-molecule activation mechanism that directly opens the KCNQ2 channel.
Nat.Chem.Biol., 20, 2024
|
|
8I2K
 
 | |
8IHL
 
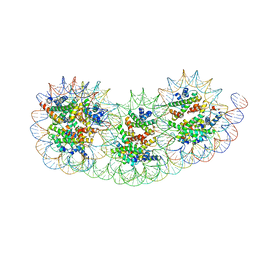 | | Overlapping tri-nucleosome | | Descriptor: | DNA (353-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Nishimura, M, Fujii, T, Tanaka, H, Maehara, K, Nozawa, K, Takizawa, Y, Ohkawa, Y, Kurumizaka, H. | | Deposit date: | 2023-02-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (7.64 Å) | | Cite: | Genome-wide mapping and cryo-EM structural analyses of the overlapping tri-nucleosome composed of hexasome-hexasome-octasome moieties.
Commun Biol, 7, 2024
|
|
8I9U
 
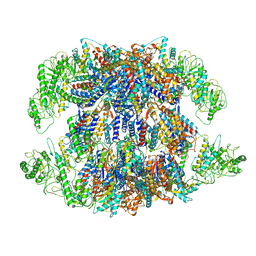 | | Human TRiC-PhLP2A complex in the open state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosducin-like protein 3, T-complex protein 1 subunit alpha, ... | | Authors: | Roh, S.H, Park, J, Kim, H, Lim, S. | | Deposit date: | 2023-02-07 | | Release date: | 2024-01-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A structural vista of phosducin-like PhLP2A-chaperonin TRiC cooperation during the ATP-driven folding cycle.
Nat Commun, 15, 2024
|
|
8I1U
 
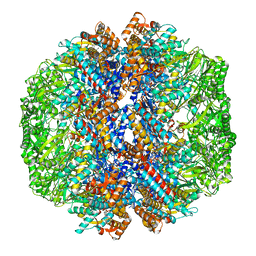 | | Human TRiC-PhLP2A complex in the closed state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Roh, S.H, Park, J, Kim, H, Lim, S. | | Deposit date: | 2023-01-13 | | Release date: | 2024-01-31 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | A structural vista of phosducin-like PhLP2A-chaperonin TRiC cooperation during the ATP-driven folding cycle.
Nat Commun, 15, 2024
|
|
