6O2W
 
 | |
6HKU
 
 | |
8SIC
 
 | | Crystal structure of Epstein-Barr virus glycoprotein 350 (gp350) in complex with Cy137C02, a monoclonal antibody isolated from macaques immunized with a gp350 nanoparticle vaccine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cy137C02 Fab heavy chain, Cy137C02 Fab light chain, ... | | Authors: | Joyce, M.G, Jensen, J.L, Chen, W.H, Kanekiyo, M. | | Deposit date: | 2023-04-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Crystal structure of Epstein-Barr virus glycoprotein 350 (gp350) in complex with Cy137C02, a monoclonal antibody isolated from macaques immunized with a gp350 nanoparticle vaccine
To Be Published
|
|
6EJ7
 
 | |
6NVT
 
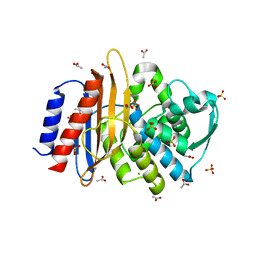 | | Crystal structure of TLA-1 extended spectrum Beta-lactamase | | Descriptor: | ACETATE ION, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Rudino-Pinera, E, Cifuentes-Castro, V.H, Rodriguez-Almazan, C. | | Deposit date: | 2019-02-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of ESBL TLA-1 in complex with clavulanic acid reveals a second acylation site.
Biochem.Biophys.Res.Commun., 522, 2020
|
|
8JVA
 
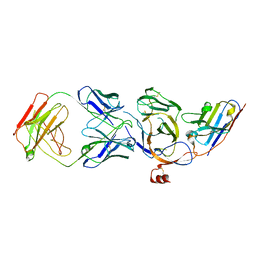 | | Cryo-EM structure of the N-terminal domain of Omicron BA.1 in complex with nanobody N235 and S2L20 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S2L20 heavy chain, S2L20 light chain, ... | | Authors: | Liu, B, Liu, H.H, Han, P, Qi, J.X. | | Deposit date: | 2023-06-28 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Enhanced potency of an IgM-like nanobody targeting conserved epitope in SARS-CoV-2 spike N-terminal domain.
Signal Transduct Target Ther, 9, 2024
|
|
6NYW
 
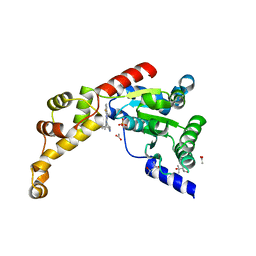 | | Structure of spastin AAA domain N527C mutant in complex with 8-fluoroquinazoline-based inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, N-(5-tert-butyl-1H-pyrazol-3-yl)-8-fluoro-2-[(2R)-2-methylpiperazin-1-yl]quinazolin-4-amine, ... | | Authors: | Pisa, R, Cupido, T, Kapoor, T.M. | | Deposit date: | 2019-02-12 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Designing Allele-Specific Inhibitors of Spastin, a Microtubule-Severing AAA Protein.
J. Am. Chem. Soc., 141, 2019
|
|
6NZQ
 
 | |
6NW5
 
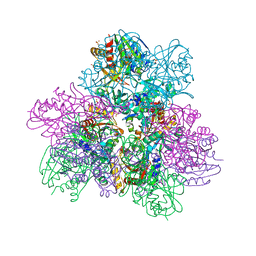 | |
8K5O
 
 | | Cryo-EM structure of the RC-LH core comples from Halorhodospira halochloris | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 2-O-octyl-beta-D-glucopyranose, ... | | Authors: | Wang, G.-L, Qi, C.-H, Yu, L.-J. | | Deposit date: | 2023-07-22 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Structural insights into the unusual core photocomplex from a triply extremophilic purple bacterium, Halorhodospira halochloris.
J Integr Plant Biol, 2024
|
|
5K9O
 
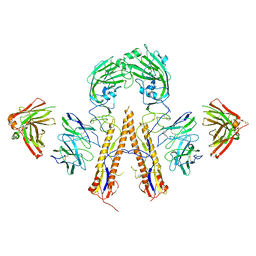 | | Crystal structure of multidonor HV1-18+HD3-9 class broadly neutralizing Influenza A antibody 31.b.09 in complex with Hemagglutinin H1 A/California/04/2009 | | Descriptor: | 31.b.09 Heavy Fv, 31.b.09 Light Fv, Hemagglutinin, ... | | Authors: | Joyce, M.G, Thomas, P.V, Wheatley, A.K, McDermott, A.B, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-06-01 | | Release date: | 2016-12-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.387 Å) | | Cite: | Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses.
Cell, 166, 2016
|
|
6NZF
 
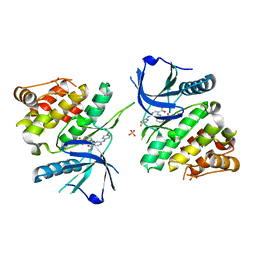 | |
6YSX
 
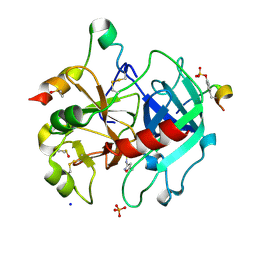 | | Thrombin in complex with 4-amino-N-(5-methylisoxazol-3-yl)benzenesulfonamide (j80) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, Hirudin variant-2, ... | | Authors: | Scanlan, W, Heine, A, Klebe, G, Abazi, N. | | Deposit date: | 2020-04-23 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Thrombin in complex with 4-amino-N-(5-methylisoxazol-3-yl)benzenesulfonamide (j80)
To be published
|
|
6YWA
 
 | |
6YWB
 
 | |
6NXS
 
 | | Crystal structure of Arabidopsis thaliana cytosolic triosephosphate isomerase C218Y mutant | | Descriptor: | SODIUM ION, Triosephosphate isomerase, cytosolic | | Authors: | Jimenez-Sandoval, P, Diaz-Quezada, C, Torres-Larios, A, Brieba, L.G. | | Deposit date: | 2019-02-09 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural basis for the modulation of plant cytosolic triosephosphate isomerase activity by mimicry of redox-based modifications.
Plant J., 99, 2019
|
|
6YSJ
 
 | | Thrombin in complex with 2-amino-1-(4-bromophenyl)ethan-1-one (j10) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-1-(4-bromophenyl)ethanone, DIMETHYL SULFOXIDE, ... | | Authors: | Scanlan, W, Heine, A, Klebe, G, Abazi, N, Paulus, A. | | Deposit date: | 2020-04-22 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Thrombin in complex with 2-amino-1-(4-bromophenyl)ethan-1-one (j10)
To be published
|
|
6NZE
 
 | |
6NZP
 
 | |
8S7C
 
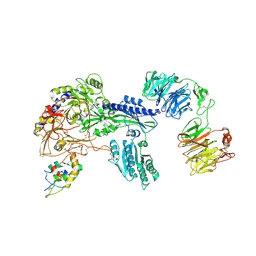 | | Ternary Complex of Cachd1, FZD5 and LRP6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Frizzled-5, ... | | Authors: | Zhao, Y, Ren, J, Jones, E.Y. | | Deposit date: | 2024-02-29 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (4.7 Å) | | Cite: | Cachd1 interacts with Wnt receptors and regulates neuronal asymmetry in the zebrafish brain.
Science, 384, 2024
|
|
6Z0Z
 
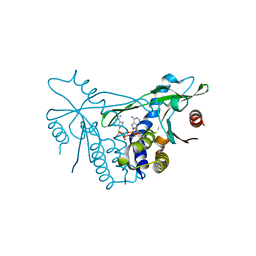 | | Human wtSTING in complex with 3',3'-c-(2'FdAMP-2'FdAMP) | | Descriptor: | 2'-fluoro-,3',3'-c-di-AMP, Stimulator of interferon protein | | Authors: | Boura, E, Smola, M. | | Deposit date: | 2020-05-11 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Ligand Strain and Its Conformational Complexity Is a Major Factor in the Binding of Cyclic Dinucleotides to STING Protein.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6O0N
 
 | | M.tb MenD with Inhibitor | | Descriptor: | 1,4-dihydroxy-2-naphthoic acid, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase | | Authors: | Johnston, J.M, Ho, N.A.T, Bashiri, G, Bulloch, E.M, Nigon, L.V, Jirgis, E.M.N, Baker, E.N. | | Deposit date: | 2019-02-16 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Allosteric regulation of menaquinone (vitamin K2) biosynthesis in the human pathogenMycobacterium tuberculosis.
J.Biol.Chem., 295, 2020
|
|
8S0L
 
 | | Crystal structure of the TMPRSS2 zymogen in complex with the nanobody A07 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Nanobody A07, ... | | Authors: | Duquerroy, S, Fernandez, I, Rey, F. | | Deposit date: | 2024-02-14 | | Release date: | 2024-06-26 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of TMPRSS2 zymogen activation and recognition by the HKU1 seasonal coronavirus.
Cell, 187, 2024
|
|
6O2V
 
 | |
7R85
 
 | | Structure of mouse Bai1 (ADGRB1) TSR3 domain | | Descriptor: | Vasculostatin-120, alpha-D-mannopyranose, beta-D-glucopyranose-(1-3)-alpha-L-fucopyranose | | Authors: | Miao, Y, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-06-26 | | Release date: | 2021-11-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | RTN4/NoGo-receptor binding to BAI adhesion-GPCRs regulates neuronal development.
Cell, 184, 2021
|
|
