5NYW
 
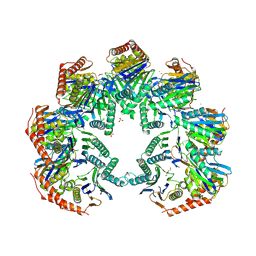 | | Anbu (ancestral beta-subunit) from Yersinia bercovieri | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, CHLORIDE ION, ... | | Authors: | Piasecka, A, Czapinska, H, Vielberg, M, Szczepanowski, R.H, Reed, S, Groll, M, Bochtler, M. | | Deposit date: | 2017-05-12 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | The Y. bercovieri Anbu crystal structure sheds light on the evolution of highly (pseudo)symmetric multimers.
J. Mol. Biol., 430, 2018
|
|
5OBD
 
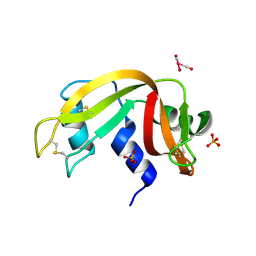 | | X-ray structure of the adduct formed upon reaction of ribonuclease A with the compound fac-[RuII(CO)3Cl2(N3-MIM), MIM=methyl-imidazole | | Descriptor: | PHOSPHATE ION, Ribonuclease pancreatic, tris(oxidaniumylidynemethyl)-tris(oxidanyl)ruthenium | | Authors: | Pontillo, N, Ferraro, G, Merlino, A. | | Deposit date: | 2017-06-26 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Ru-Based CO releasing molecules with azole ligands: interaction with proteins and the CO release mechanism disclosed by X-ray crystallography.
Dalton Trans, 46, 2017
|
|
6PEW
 
 | |
5O11
 
 | |
6P7P
 
 | | Structure of E. coli MS115-1 NucC, cAAA-bound form | | Descriptor: | CHLORIDE ION, Cyclic tri-AMP (5'-3' linked), E. coli MS115-1 NucC 2-241 | | Authors: | Ye, Q, Lau, R.K, Berg, K.R, Corbett, K.D. | | Deposit date: | 2019-06-06 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | Structure and Mechanism of a Cyclic Trinucleotide-Activated Bacterial Endonuclease Mediating Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6PB6
 
 | |
6P85
 
 | | E.coli LpxD in complex with compound 3 | | Descriptor: | 1-(2-ethoxyphenyl)-4-[(4-methylphenyl)sulfonyl]-1H-1,2,3-triazol-5-amine, 3-hydroxy-7,7-dimethyl-2-phenyl-4-(thiophen-2-yl)-2,6,7,8-tetrahydro-5H-pyrazolo[3,4-b]quinolin-5-one, MAGNESIUM ION, ... | | Authors: | Ma, X, Shia, S. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Biological Basis of Small Molecule Inhibition ofEscherichia coliLpxD Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
Acs Infect Dis., 6, 2020
|
|
6PHF
 
 | |
5OD2
 
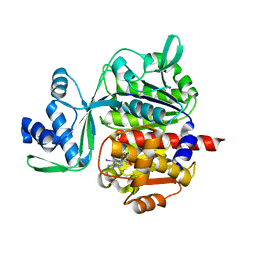 | | Crystal structure of ADP-dependent glucokinase from Methanocaldococcus jannaschii | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, Bifunctional ADP-specific glucokinase/phosphofructokinase, MAGNESIUM ION, ... | | Authors: | Wisniewska, M, Tokarz, P, Grudnik, P. | | Deposit date: | 2017-07-04 | | Release date: | 2018-01-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of ADP-dependent glucokinase from Methanocaldococcus jannaschii in complex with 5-iodotubercidin reveals phosphoryl transfer mechanism.
Protein Sci., 27, 2018
|
|
6PO3
 
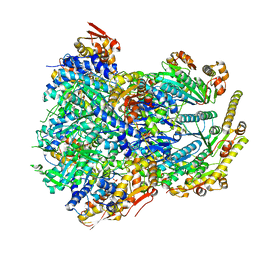 | | ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | Deposit date: | 2019-07-03 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.28 Å) | | Cite: | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
7WJZ
 
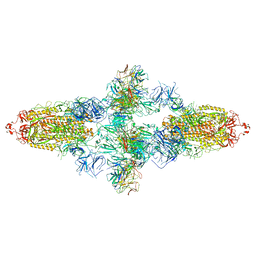 | | Omicron Spike bitrimer with 6m6 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6m6 heavy chain, 6m6 light chain, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-08 | | Release date: | 2022-07-13 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | A broadly neutralizing antibody against SARS-CoV-2 Omicron variant infection exhibiting a novel trimer dimer conformation in spike protein binding.
Cell Res., 32, 2022
|
|
5ODM
 
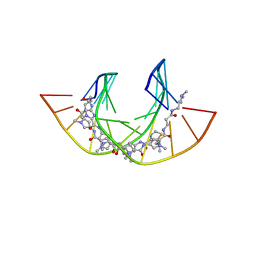 | | NtiPr polyamide in complex with 5'CGATGTACTACG3 | | Descriptor: | 3-(3-azaniumylpropanoylamino)propyl-dimethyl-azanium, 4-AMINO-(1-METHYLIMIDAZOLE)-2-CARBOXYLIC ACID, 4-AMINO-(1-METHYLPYRROLE)-2-CARBOXYLIC ACID, ... | | Authors: | Padroni, G, Parkinson, J, Burley, G.A. | | Deposit date: | 2017-07-06 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of DNA duplex distortion induced by thiazole-containing hairpin polyamides.
Nucleic Acids Res., 46, 2018
|
|
6P9Q
 
 | | E.coli LpxA in complex with UDP-3-O-(R-3-hydroxymyristoyl)-GlcNAc and Compound 2 | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, DIMETHYL SULFOXIDE, PHOSPHATE ION, ... | | Authors: | Ma, X, Shia, S, Ornelas, E. | | Deposit date: | 2019-06-10 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Two Distinct Mechanisms of Inhibition of LpxA Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
J.Am.Chem.Soc., 142, 2020
|
|
5OE2
 
 | | CRYSTAL STRUCTURE OF THE BETA-LACTAMASE OXA-245 | | Descriptor: | Beta-lactamase, CHLORIDE ION, SODIUM ION | | Authors: | Lund, B.A, Leiros, H.K.S. | | Deposit date: | 2017-07-07 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure, activity and thermostability investigations of OXA-163, OXA-181 and OXA-245 using biochemical analysis, crystal structures and differential scanning calorimetry analysis.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5NV4
 
 | | UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum double mutant D611C:G1050C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FORMIC ACID, UDP-glucose-glycoprotein glucosyltransferase-like protein, ... | | Authors: | Roversi, P, Caputo, A.T, Hill, J, Alonzi, D.S, Zitzmann, N. | | Deposit date: | 2017-05-03 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Interdomain conformational flexibility underpins the activity of UGGT, the eukaryotic glycoprotein secretion checkpoint.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7WJV
 
 | | Crystal structure of human liver FBPase complexed with an covalent inhibitor | | Descriptor: | 1,2-BENZISOTHIAZOL-3(2H)-ONE 1,1-DIOXIDE, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Cao, H, Huang, Y, Ren, Y, Wan, J. | | Deposit date: | 2022-01-08 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.724 Å) | | Cite: | N -Acylamino Saccharin as an Emerging Cysteine-Directed Covalent Warhead and Its Application in the Identification of Novel FBPase Inhibitors toward Glucose Reduction.
J.Med.Chem., 65, 2022
|
|
5NW2
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-3,3-dimethyl-2-(oxetane-3-carboxamido)butanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 19) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-3,3-dimethyl-2-(oxetan-3-ylcarbonylamino)butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Gadd, M.S, Soares, P, Ciulli, A. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Group-Based Optimization of Potent and Cell-Active Inhibitors of the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase: Structure-Activity Relationships Leading to the Chemical Probe (2S,4R)-1-((S)-2-(1-Cyanocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (VH298).
J. Med. Chem., 61, 2018
|
|
6PC9
 
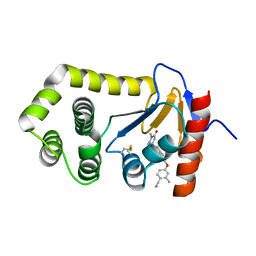 | | Crystal Structure of EcDsbA in a complex with purified methylpiperazinone 6 | | Descriptor: | 2-methyl-4-{4-[2-(4-methyl-3-oxopiperazin-1-yl)-2-oxoethyl]phenoxy}benzonitrile, TRIETHYLENE GLYCOL, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-17 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiLX).
J.Med.Chem., 63, 2020
|
|
6PNH
 
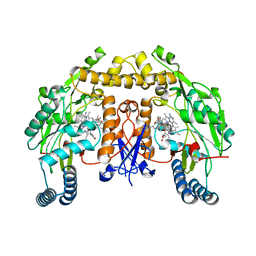 | |
6POA
 
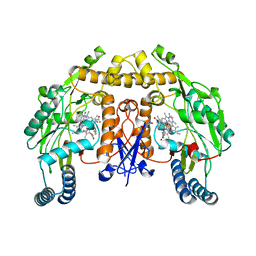 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 7-(3-(Aminomethyl)-4-(pyridin-3-ylmethoxy)phenyl)-4-methylquinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-{3-(aminomethyl)-4-[(pyridin-3-yl)methoxy]phenyl}-4-methylquinolin-2-amine, Nitric oxide synthase, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2019-07-03 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.809 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
6PEI
 
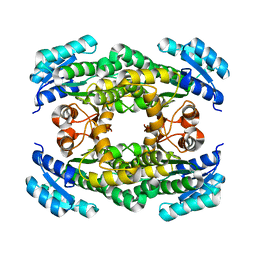 | | Structure of sorbitol dehydrogenase from Sinorhizobium meliloti 1021 | | Descriptor: | Sorbitol dehydrogenase (L-iditol 2-dehydrogenase) | | Authors: | Bailey-Elkin, B.A, Kohlmeier, M.G, Oresnik, I.J, Mark, B.L. | | Deposit date: | 2019-06-20 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of the sorbitol dehydrogenase SmoS from Sinorhizobium meliloti 1021
Acta Crystallogr.,Sect.D, 77, 2021
|
|
5OGX
 
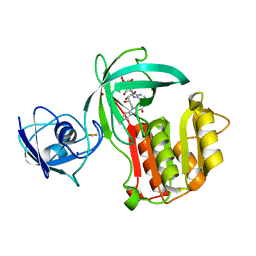 | | Crystal structure of Amycolatopsis cytochrome P450 reductase GcoB. | | Descriptor: | BROMIDE ION, Cytochrome P450 reductase, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Mallinson, S.J.B, Johnson, C.W, Neidle, E.L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-07-13 | | Release date: | 2018-07-04 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion.
Nat Commun, 9, 2018
|
|
6POH
 
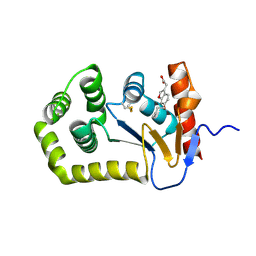 | | Crystal Structure of EcDsbA in complex alkyl ether 21 | | Descriptor: | (6-butoxy-1-benzofuran-3-yl)acetic acid, COPPER (II) ION, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Scanlon, M.J. | | Deposit date: | 2019-07-03 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The Fragment-Based Development of a Benzofuran Hit as a New Class of Escherichia coli DsbA Inhibitors.
Molecules, 24, 2019
|
|
6P8U
 
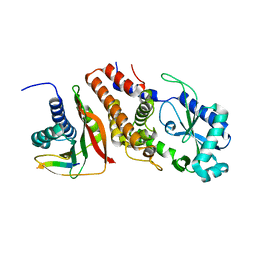 | | Structure of P. aeruginosa ATCC27853 CdnD:HORMA2:Peptide 1 complex | | Descriptor: | HORMA domain containing protein, MAGNESIUM ION, Nucleotidyltransferase, ... | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-06-08 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6PFB
 
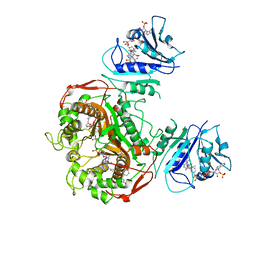 | | Crystal structure of TS-DHFR from Cryptosporidium hominis in complex with NADPH, FdUMP and 3-(2-(4-((2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl)benzamido)phenyl)propanoic acid. | | Descriptor: | 3-[2-({4-[(2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl]benzene-1-carbonyl}amino)phenyl]propanoic acid, 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Czyzyk, D.J, Valhondo, M, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2019-06-21 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.089 Å) | | Cite: | Structure activity relationship towards design of cryptosporidium specific thymidylate synthase inhibitors.
Eur.J.Med.Chem., 183, 2019
|
|
