6ONV
 
 | | Crystal structure of HIV-1 LM/HT Clade A/E CRF01 gp120 core in complex with (S)-MCG-III-027-D05. | | Descriptor: | (3S)-N-(4-chloro-3-fluorophenyl)-1-(methylsulfonyl)piperidine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tolbert, W.D, Sherburn, R, Pazgier, M. | | Deposit date: | 2019-04-22 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.253 Å) | | Cite: | A New Family of Small-Molecule CD4-Mimetic Compounds Contacts Highly Conserved Aspartic Acid 368 of HIV-1 gp120 and Mediates Antibody-Dependent Cellular Cytotoxicity.
J.Virol., 93, 2019
|
|
6ONF
 
 | | Crystal structure of HIV-1 LM/HT Clade A/E CRF01 gp120 core in complex with (S)-MCG-III-188-A02. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, clade A/E 93TH057 HIV-1 gp120 core, ... | | Authors: | Tolbert, W.D, Sherburn, R, Pazgier, M. | | Deposit date: | 2019-04-22 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | A New Family of Small-Molecule CD4-Mimetic Compounds Contacts Highly Conserved Aspartic Acid 368 of HIV-1 gp120 and Mediates Antibody-Dependent Cellular Cytotoxicity.
J.Virol., 93, 2019
|
|
6E36
 
 | |
6E3F
 
 | |
6BQ2
 
 | |
6BQ5
 
 | | Crystal structure of Medicago truncatula Thermospermine Synthase (MtTSPS) in complex with 5'-methylthioadenosine | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, GLYCEROL, ... | | Authors: | Sekula, B, Dauter, Z. | | Deposit date: | 2017-11-27 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of thermospermine synthase fromMedicago truncatulaand substrate discriminatory features of plant aminopropyltransferases.
Biochem. J., 475, 2018
|
|
6BQ4
 
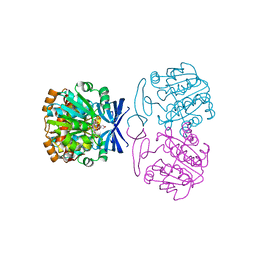 | | Crystal structure of Medicago truncatula Thermospermine Synthase (MtTSPS) in complex with adenosine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE, ... | | Authors: | Sekula, B, Dauter, Z. | | Deposit date: | 2017-11-27 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of thermospermine synthase fromMedicago truncatulaand substrate discriminatory features of plant aminopropyltransferases.
Biochem. J., 475, 2018
|
|
6CD1
 
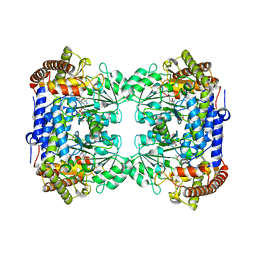 | | Crystal structure of Medicago truncatula serine hydroxymethyltransferase 3 (MtSHMT3), complexes with reaction intermediates | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GLYCINE, ... | | Authors: | Ruszkowski, M, Sekula, B, Ruszkowska, A, Dauter, Z. | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Chloroplastic Serine Hydroxymethyltransferase FromMedicago truncatula: A Structural Characterization.
Front Plant Sci, 9, 2018
|
|
5ICY
 
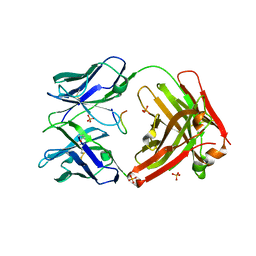 | | Cetuximab Fab in complex with linear meditope | | Descriptor: | Cetuximab Fab heavy chain, Cetuximab Fab light chain, Meditope, ... | | Authors: | Bzymek, K.P, Williams, J.C. | | Deposit date: | 2016-02-23 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cyclization strategies of meditopes: affinity and diffraction studies of meditope-Fab complexes.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
5IOP
 
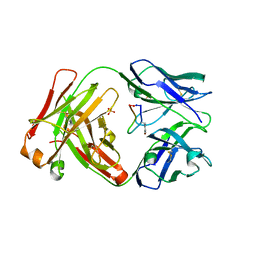 | |
5IV2
 
 | | Cetuximab Fab in complex with Arg9Cir meditope variant | | Descriptor: | Cetuximab Fab, heavy chain, light chain, ... | | Authors: | Bzymek, K.P, Williams, J.C. | | Deposit date: | 2016-03-18 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.481 Å) | | Cite: | Natural and non-natural amino-acid side-chain substitutions: affinity and diffraction studies of meditope-Fab complexes.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
5IVZ
 
 | | Cetuximab Fab in complex with Arg8Cir meditope variant | | Descriptor: | Cetuximab Fab, heavy chain, light chain, ... | | Authors: | Bzymek, K.P, Williams, J.C. | | Deposit date: | 2016-03-21 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Natural and non-natural amino-acid side-chain substitutions: affinity and diffraction studies of meditope-Fab complexes.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
2MTE
 
 | | Solution structure of Doc48S | | Descriptor: | CALCIUM ION, Cellulose 1,4-beta-cellobiosidase (reducing end) CelS | | Authors: | Chen, C, Feng, Y. | | Deposit date: | 2014-08-18 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Revisiting the NMR solution structure of the Cel48S type-I dockerin module from Clostridium thermocellum reveals a cohesin-primed conformation.
J.Struct.Biol., 188, 2014
|
|
5FLY
 
 | | The FhuD protein from S.pseudintermedius | | Descriptor: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Malito, E. | | Deposit date: | 2015-10-29 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Crystal Structure of Fhud at 1.6 Angstrom Resolution: A Ferrichrome-Binding Protein from the Animal and Human Pathogen Staphylococcus Pseudintermedius
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5ETU
 
 | | Cetuximab Fab in complex with L5E meditope variant | | Descriptor: | Cetuximab Fab heavy chain, Cetuximab Fab light chain, L5E meditope variant, ... | | Authors: | Bzymek, K.P, Williams, J.C. | | Deposit date: | 2015-11-18 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Natural and non-natural amino-acid side-chain substitutions: affinity and diffraction studies of meditope-Fab complexes.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
5ITF
 
 | |
5TH2
 
 | | Cetuximab Fab in complex with L5Q meditope variant | | Descriptor: | L5Q meditope, PHOSPHATE ION, cetuximab Fab, ... | | Authors: | Bzymek, K.P, Williams, J.C. | | Deposit date: | 2016-09-28 | | Release date: | 2016-10-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Natural and non-natural amino-acid side-chain substitutions: affinity and diffraction studies of meditope-Fab complexes.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
4OXR
 
 | | Structure of Staphylococcus pseudintermedius metal-binding protein SitA in complex with Manganese | | Descriptor: | MANGANESE (II) ION, Manganese ABC transporter, periplasmic-binding protein SitA | | Authors: | Abate, F, Malito, E, Bottomley, M. | | Deposit date: | 2014-02-06 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Apo, Zn2+-bound and Mn2+-bound structures reveal ligand-binding properties of SitA from the pathogen Staphylococcus pseudintermedius.
Biosci.Rep., 34, 2014
|
|
4OXQ
 
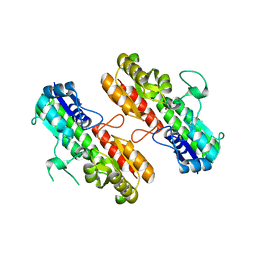 | | Structure of Staphylococcus pseudintermedius metal-binding protein SitA in complex with Zinc | | Descriptor: | Manganese ABC transporter, periplasmic-binding protein SitA, ZINC ION | | Authors: | Abate, F, Malito, E, Bottomley, M. | | Deposit date: | 2014-02-06 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Apo, Zn2+-bound and Mn2+-bound structures reveal ligand-binding properties of SitA from the pathogen Staphylococcus pseudintermedius.
Biosci.Rep., 34, 2014
|
|
1P3C
 
 | | Glutamyl endopeptidase from Bacillus intermedius | | Descriptor: | glutamyl-endopeptidase | | Authors: | Meijers, R, Blagova, E.V, Levdikov, V.M, Rudenskaya, G.N, Chestukhina, G.G, Akimkina, T.V, Kostrov, S.V, Lamzin, V.S, Kuranova, I.P. | | Deposit date: | 2003-04-17 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of glutamyl endopeptidase from Bacillus intermedius reveals a structural link between zymogen activation and charge compensation.
Biochemistry, 43, 2004
|
|
4IS4
 
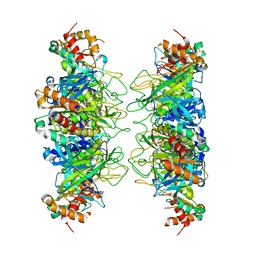 | |
3BB7
 
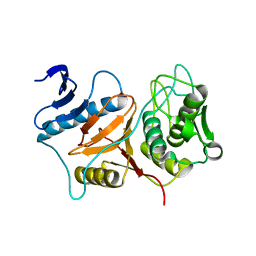 | | Structure of Prevotella intermedia prointerpain A fragment 39-359 (mutant C154A) | | Descriptor: | interpain A | | Authors: | Mallorqui-Fernandez, N, Manandhar, S.P, Mallorqui-Fernandez, G, Uson, I, Wawrzonek, K, Kantyka, T, Sola, M, Thogersen, I.B, Enghild, J.J, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2007-11-09 | | Release date: | 2007-11-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A New Autocatalytic Activation Mechanism for Cysteine Proteases Revealed by Prevotella intermedia Interpain A
J.Biol.Chem., 283, 2008
|
|
2NOR
 
 | |
4UWH
 
 | | Discovery of (2S)-8-((3R)-3-Methylmorpholin-4-yl)-1-(3-methyl-2-oxo- butyl)-2-(trifluoromethyl)-3,4-dihydro-2H-pyrimido(1,2-a)pyrimidin-6- one: a Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors | | Descriptor: | (8S)-9-[(2R)-2-hydroxy-2-phenylethyl]-2-(morpholin-4-yl)-8-(trifluoromethyl)-6,7,8,9-tetrahydro-4H-pyrimido[1,2-a]pyrimidin-4-one, PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT TYPE 3, SODIUM ION | | Authors: | Pasquier, B, El-Ahmad, Y, Filoche-Romme, B, Dureuil, C, Fassy, F, Abecassis, P.Y, Mathieu, M, Bertrand, T, Benard, T, Barriere, C, ElBatti, S, Letallec, J.P, Sonnefraud, V, Brollo, M, Delbarre, L, Loyau, V, Pilorge, F, Bertin, L, Richepin, P, Arigon, J, Labrosse, J.R, Clement, J, Durand, F, Combet, R, Perraut, P, Leroy, V, Gay, F, Lefrancois, D, Bretin, F, Marquette, J.P, Michot, N, Caron, A, Castell, C, Schio, L, McCort, G, Goulaouic, H, Garcia-Echeverria, C, Ronan, B. | | Deposit date: | 2014-08-12 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Discovery of (2S)-8-[(3R)-3-Methylmorpholin-4-Yl]-1-(3-Methyl-2-Oxo-Butyl)-2-(Trifluoromethyl)-3,4-Dihydro-2H-Pyrimido[1,2-A]Pyrimidin-6-One: A Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors.
J.Med.Chem., 58, 2015
|
|
2RIP
 
 | | Structure of DPPIV in complex with an inhibitor | | Descriptor: | (3R,4R)-4-(pyrrolidin-1-ylcarbonyl)-1-(quinoxalin-2-ylcarbonyl)pyrrolidin-3-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Qiu, X. | | Deposit date: | 2007-10-12 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design and synthesis of potent amido- and benzyl-substituted cis-3-amino-4-(2-cyanopyrrolidide)pyrrolidinyl DPP-IV inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
