5IGN
 
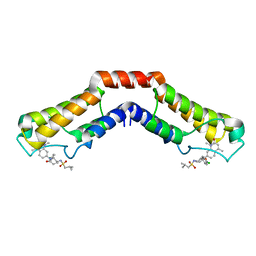 | | Crystal structure of human BRD9 bromodomain in complex with LP99 chemical probe | | Descriptor: | Bromodomain-containing protein 9, N-[(2R,3S)-2-(4-chlorophenyl)-1-(1,4-dimethyl-2-oxo-1,2-dihydroquinolin-7-yl)-6-oxopiperidin-3-yl]-2-methylpropane-1-sulfonamide | | Authors: | Tallant, C, Clark, P.G.K, Vieira, L.C.C, Newman, J.A, Krojer, T, Nunez-Alonso, G, Picaud, S, Fedorov, O, Dixon, D.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-28 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of BRD9 bromodomain in complex with LP99 chemical probe
To Be Published
|
|
2Q40
 
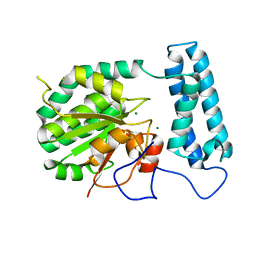 | | Ensemble refinement of the protein crystal structure of gene product from Arabidopsis thaliana At2g17340 | | Descriptor: | MAGNESIUM ION, Protein At2g17340 | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
7M1D
 
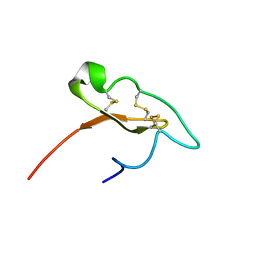 | |
7TJ4
 
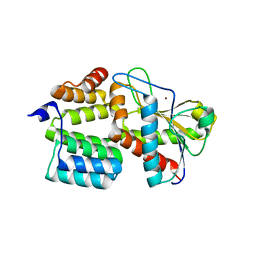 | | Structure of the S. aureus amidase LytH and activator ActH extracellular domains | | Descriptor: | ActH, LytH, ZINC ION | | Authors: | Page, J.E, Skiba, M.A, Kruse, A.C, Walker, S. | | Deposit date: | 2022-01-14 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Metal cofactor stabilization by a partner protein is a widespread strategy employed for amidase activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2C9Y
 
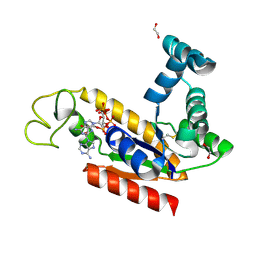 | | Structure of human adenylate kinase 2 | | Descriptor: | 1,2-ETHANEDIOL, ADENYLATE KINASE ISOENZYME 2, MITOCHONDRIAL, ... | | Authors: | Bunkoczi, G, Filippakopoulos, P, Debreczeni, J.E, Turnbull, A, Papagrigoriou, E, Savitsky, P, Colebrook, S, von Delft, F, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, Knapp, S. | | Deposit date: | 2005-12-15 | | Release date: | 2006-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Human Adenylate Kinase 2
To be Published
|
|
6GQK
 
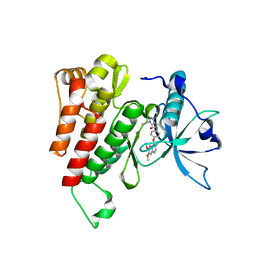 | | Crystal structure of human c-KIT kinase domain in complex with AZD3229-analogue (compound 23) | | Descriptor: | Mast/stem cell growth factor receptor Kit, ~{N}-[4-(6,7-dimethoxyquinazolin-4-yl)oxyphenyl]-2-(1-ethylpyrazol-4-yl)ethanamide | | Authors: | Schimpl, M, Hardy, C.J, Ogg, D.J, Overman, R.C, Packer, M.J, Kettle, J.G, Anjum, R, Barry, E, Bhavsar, D, Brown, C, Campbell, A, Goldberg, K, Grondine, M, Guichard, S, Hunt, T, Jones, O, Li, X, Moleva, O, Pearson, S, Shao, W, Smith, A, Smith, J, Stead, D, Stokes, S, Tucker, M, Ye, Y. | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery of N-(4-{[5-Fluoro-7-(2-methoxyethoxy)quinazolin-4-yl]amino}phenyl)-2-[4-(propan-2-yl)-1 H-1,2,3-triazol-1-yl]acetamide (AZD3229), a Potent Pan-KIT Mutant Inhibitor for the Treatment of Gastrointestinal Stromal Tumors.
J. Med. Chem., 61, 2018
|
|
7NFU
 
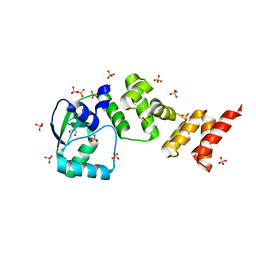 | | Crystal structure of C-terminally truncated Geobacillus thermoleovorans nucleoid occlusion protein Noc | | Descriptor: | GLYCEROL, Nucleoid occlusion protein, SULFATE ION | | Authors: | Jalal, A.S.B, Tran, N.T, Wu, L.J, Ramakrishnan, K, Rejzek, M, Stevenson, C.E.M, Lawson, D.M, Errington, J, Le, T.B.K. | | Deposit date: | 2021-02-07 | | Release date: | 2021-02-17 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | CTP regulates membrane-binding activity of the nucleoid occlusion protein Noc.
Mol.Cell, 81, 2021
|
|
8W26
 
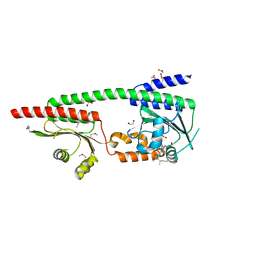 | |
6I43
 
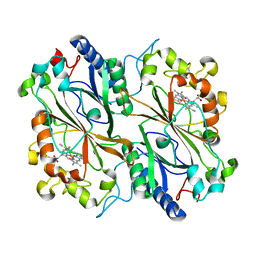 | | SFX Structure of Damage Free Ferric State of Dye Type Peroxidase Aa from Streptomyces lividans | | Descriptor: | Deferrochelatase/peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ebrahim, A, Moreno-Chicano, T, Chaplin, A.K, Sherrell, D.A, Duyvesteyn, H.M.E, Owada, S, Tono, K, Sugimoto, H, Strange, R.W, Worrall, J.A.R. | | Deposit date: | 2018-11-08 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Dose-resolved serial synchrotron and XFEL structures of radiation-sensitive metalloproteins.
Iucrj, 6, 2019
|
|
2PPD
 
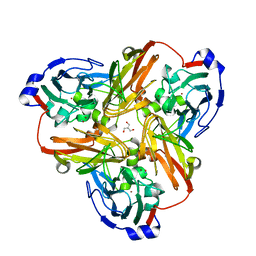 | |
3E1O
 
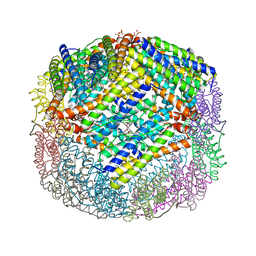 | | Crystal structure of E. coli Bacterioferritin (BFR) with two ZN(II) ION sites at the Ferroxidase centre (ZN-BFR). | | Descriptor: | BACTERIOFERRITIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Crow, A, Lawson, T, Lewin, A, Moore, G.R, Le Brun, N. | | Deposit date: | 2008-08-04 | | Release date: | 2009-05-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for iron mineralization by bacterioferritin
J.Am.Chem.Soc., 131, 2009
|
|
6I4X
 
 | | Crystal structure of SOCS2:Elongin C:Elongin B in complex with erythropoietin receptor peptide | | Descriptor: | DI(HYDROXYETHYL)ETHER, Elongin-B, Elongin-C, ... | | Authors: | Kung, W.W, Ramachandran, S, Makukhin, N, Bruno, E, Ciulli, A. | | Deposit date: | 2018-11-12 | | Release date: | 2019-05-29 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural insights into substrate recognition by the SOCS2 E3 ubiquitin ligase.
Nat Commun, 10, 2019
|
|
5VD2
 
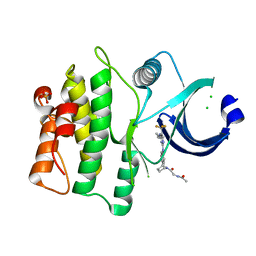 | | crystal structure of human WEE1 kinase domain in complex with PF-03814735 | | Descriptor: | CHLORIDE ION, N-{2-[(1S,4R)-6-{[4-(cyclobutylamino)-5-(trifluoromethyl)pyrimidin-2-yl]amino}-1,2,3,4-tetrahydro-1,4-epiminonaphthalen-9-yl]-2-oxoethyl}acetamide, PHOSPHATE ION, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-04-01 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis of Wee Kinases Functionality and Inactivation by Diverse Small Molecule Inhibitors.
J. Med. Chem., 60, 2017
|
|
4F21
 
 | | Crystal structure of carboxylesterase/phospholipase family protein from Francisella tularensis | | Descriptor: | Carboxylesterase/phospholipase family protein, N-((1R,2S)-2-allyl-4-oxocyclobutyl)-4-methylbenzenesulfonamide, bound form | | Authors: | Filippova, E.V, Minasov, G, Kuhn, M, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Winsor, J.R, Kiryukhina, O, Becker, D.P, Armoush, N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-07 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Large scale structural rearrangement of a serine hydrolase from Francisella tularensis facilitates catalysis.
J.Biol.Chem., 288, 2013
|
|
1OP8
 
 | | Crystal Structure of Human Granzyme A | | Descriptor: | Granzyme A, SULFATE ION | | Authors: | Hink-Schauer, C, Estebanez-Perpina, E, Bode, W, Jenne, D. | | Deposit date: | 2003-03-05 | | Release date: | 2003-07-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the apoptosis-inducing human granzyme A dimer
NAT.STRUCT.BIOL., 10, 2003
|
|
8VYX
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with k-410 | | Descriptor: | 4,4'-[(1S,4S,5R)-5-(3,4-dihydroquinoline-1(2H)-sulfonyl)-7-oxabicyclo[2.2.1]hept-2-ene-2,3-diyl]diphenol, Estrogen receptor | | Authors: | Min, C.K, Nwachukwu, J.C, Hou, Y, Russo, R.J, Papa, A, Min, J, Peng, R, Kim, S.H, Ziegler, Y, Rangarajan, E.S, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2024-02-09 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Asymmetric allostery in estrogen receptor-alpha homodimers drives responses to the ensemble of estrogens in the hormonal milieu.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8W03
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with k-1154 | | Descriptor: | (1S,2R,4S)-N-(4-chlorophenyl)-5,6-bis(4-hydroxyphenyl)-N-(2,2,2-trifluoroethyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor | | Authors: | Min, C.K, Nwachukwu, J.C, Hou, Y, Russo, R.J, Papa, A, Min, J, Peng, R, Kim, S.H, Ziegler, Y, Rangarajan, E.S, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2024-02-13 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Asymmetric allostery in estrogen receptor-alpha homodimers drives responses to the ensemble of estrogens in the hormonal milieu.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8W07
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with k-402 | | Descriptor: | (1R,2S,4R)-N-cyclohexyl-5,6-bis(4-hydroxyphenyl)-N-(4-methoxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, (1S,2R,4S)-N-cyclohexyl-5,6-bis(4-hydroxyphenyl)-N-(4-methoxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor | | Authors: | Min, C.K, Nwachukwu, J.C, Hou, Y, Russo, R.J, Papa, A, Min, J, Peng, R, Kim, S.H, Ziegler, Y, Rangarajan, E.S, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.K, Nettles, K.W. | | Deposit date: | 2024-02-13 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Asymmetric allostery in estrogen receptor-alpha homodimers drives responses to the ensemble of estrogens in the hormonal milieu.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6UT2
 
 | | 3D structure of the leiomodin/tropomyosin binding interface | | Descriptor: | Leiomodin-2, Tropomyosin alpha-1 chain chimeric peptide | | Authors: | Tolkatchev, D, Smith, G.E, Helms, G.L, Cort, J.R, Kostyukova, A.S. | | Deposit date: | 2019-10-29 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Leiomodin creates a leaky cap at the pointed end of actin-thin filaments.
Plos Biol., 18, 2020
|
|
1FS1
 
 | | INSIGHTS INTO SCF UBIQUITIN LIGASES FROM THE STRUCTURE OF THE SKP1-SKP2 COMPLEX | | Descriptor: | CYCLIN A/CDK2-ASSOCIATED P19, CYCLIN A/CDK2-ASSOCIATED P45 | | Authors: | Schulman, B.A, Carrano, A.C, Jeffrey, P.D, Bowen, Z, Kinnucan, E.R.E, Finnin, M.S, Elledge, S.J, Harper, J.W, Pagano, M, Pavletich, N.P. | | Deposit date: | 2000-09-08 | | Release date: | 2000-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex.
Nature, 408, 2000
|
|
5MSK
 
 | | Mouse PA28beta | | Descriptor: | Proteasome activator complex subunit 2 | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2017-01-05 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The Mammalian Proteasome Activator PA28 Forms an Asymmetric alpha 4 beta 3 Complex.
Structure, 25, 2017
|
|
5IT7
 
 | | Structure of the Kluyveromyces lactis 80S ribosome in complex with the cricket paralysis virus IRES and eEF2 | | Descriptor: | (2S)-1-amino-N,N,N-trimethyl-1-oxobutan-2-aminium, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Murray, J, Savva, C.G, Shin, B.S, Dever, T.E, Ramakrishnan, V, Fernandez, I.S. | | Deposit date: | 2016-03-16 | | Release date: | 2016-05-18 | | Last modified: | 2018-11-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural characterization of ribosome recruitment and translocation by type IV IRES.
Elife, 5, 2016
|
|
4JQG
 
 | | Crystal structure of an inactive mutant of MMP-9 catalytic domain in complex with a fluorogenic synthetic peptidic substrate with a fluorine atom. | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, CALCIUM ION, ... | | Authors: | Stura, E.A, Vera, L, Cassar-Lajeunesse, E, Tranchant, I, Amoura, M, Dive, V. | | Deposit date: | 2013-03-20 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Halogen Bonding Controls Selectivity of FRET Substrate Probes for MMP-9.
Chem.Biol., 21, 2014
|
|
8TQM
 
 | |
2HFO
 
 | | Crystal Structures of the Synechocystis Photoreceptor Slr1694 Reveal Distinct Structural States Related to Signaling | | Descriptor: | Activator of photopigment and puc expression, FLAVIN MONONUCLEOTIDE | | Authors: | Yuan, H, Anderson, S, Masuda, S, Dragnea, V, Moffat, K, Bauer, C.E. | | Deposit date: | 2006-06-24 | | Release date: | 2006-12-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the Synechocystis photoreceptor Slr1694 reveal distinct structural states related to signaling.
Biochemistry, 45, 2006
|
|
