3GP7
 
 | |
3GY0
 
 | | Crystal structure of putatitve short chain dehydrogenase FROM SHIGELLA FLEXNERI 2A STR. 301 complexed with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, dehydrogenase | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-03 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of putatitve short chain dehydrogenase
FROM SHIGELLA FLEXNERI 2A STR. 301 complexed with NADP
To be Published
|
|
3GYA
 
 | |
3GPU
 
 | | MutM encountering an intrahelical 8-oxoguanine (oxoG) lesion in EC4-loop deletion complex | | Descriptor: | DNA (5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*TP*CP*GP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*CP*(8OG)P*AP*GP*TP*CP*TP*AP*CP*C)-3'), DNA glycosylase, ... | | Authors: | Banerjee, A, Qi, Y, Verdine, G.L. | | Deposit date: | 2009-03-23 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme.
Nature, 462, 2009
|
|
3GZ6
 
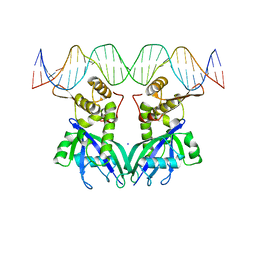 | |
3GQ3
 
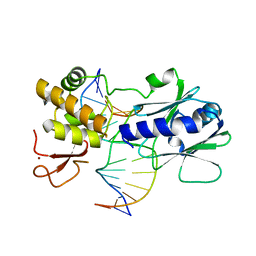 | | MutM encountering an intrahelical 8-oxoguanine (oxoG) lesion in EC5-loop deletion complex | | Descriptor: | DNA (5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*CP*CP*GP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*CP*GP*(8OG)P*GP*TP*CP*TP*AP*CP*C)-3'), DNA glycosylase, ... | | Authors: | Banerjee, A, Qi, Y, Verdine, G.L. | | Deposit date: | 2009-03-23 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme.
Nature, 462, 2009
|
|
3H14
 
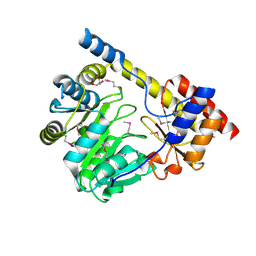 | | Crystal structure of a putative aminotransferase from Silicibacter pomeroyi | | Descriptor: | Aminotransferase, classes I and II, GLYCEROL | | Authors: | Sampathkumar, P, Atwell, S, Wasserman, S, Miller, S, Bain, K, Rutter, M, Tarun, G, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-10 | | Release date: | 2009-05-05 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a putative aminotransferase from Silicibacter pomeroyi
TO BE PUBLISHED
|
|
3H3G
 
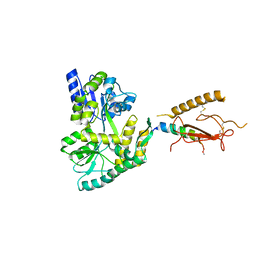 | |
3H3Q
 
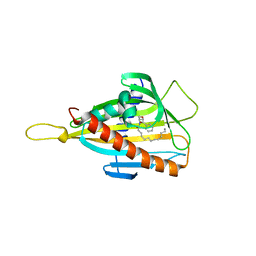 | | Crystal structure of the CERT START domain in complex with HPA-13 | | Descriptor: | Goodpasture antigen binding protein, N-[(1R,3R)-3-hydroxy-1-(hydroxymethyl)-3-phenylpropyl]tridecanamide | | Authors: | Kudo, N, Wakatsuki, S, Kato, R. | | Deposit date: | 2009-04-17 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the CERT START domain with inhibitors provide insights into the mechanism of ceramide transfer.
J.Mol.Biol., 396, 2010
|
|
3H42
 
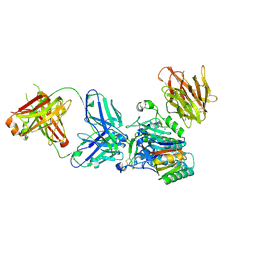 | | Crystal structure of PCSK9 in complex with Fab from LDLR competitive antibody | | Descriptor: | Fab from LDLR competitive antibody: Heavy chain, Fab from LDLR competitive antibody: Light chain, Proprotein convertase subtilisin/kexin type 9, ... | | Authors: | Piper, D.E, Walker, N.P.C, Romanow, W.G, Thibault, S.T, Tsai, M.M, Yang, E. | | Deposit date: | 2009-04-17 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | From the Cover: A proprotein convertase subtilisin/kexin type 9 neutralizing antibody reduces serum cholesterol in mice and nonhuman primates.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3H5L
 
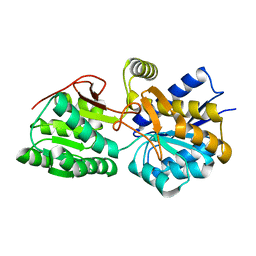 | | Crystal structure of a putative branched-chain amino acid ABC transporter from Silicibacter pomeroyi | | Descriptor: | putative Branched-chain amino acid ABC transporter | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Iizuka, M, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-22 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a putative branched-chain amino acid ABC transporter from Silicibacter pomeroyi
To be Published
|
|
3H65
 
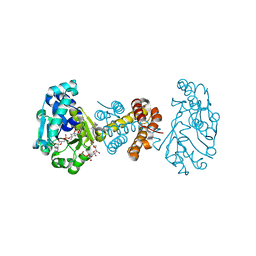 | | The Crystal Structure of C176A Mutated [Fe]-Hydrogenase (Hmd) Holoenzyme in Complex with Methylenetetrahydromethanopterin | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 5'-O-[(S)-hydroxy{[2-hydroxy-3,5-dimethyl-6-(2-oxoethyl)pyridin-4-yl]oxy}phosphoryl]guanosine, 5,10-DIMETHYLENE TETRAHYDROMETHANOPTERIN, ... | | Authors: | Hiromoto, T, Warkentin, E, Shima, S, Ermler, U. | | Deposit date: | 2009-04-23 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of an [Fe]-hydrogenase-substrate complex reveals the framework for H2 activation.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3H6D
 
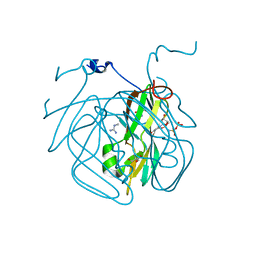 | | Structure of the mycobacterium tuberculosis DUTPase D28N mutant | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Leveles, I, Harmat, V, Nagy, G, Takacs, E, Lopata, A, Toth, J, Vertessy, B.G. | | Deposit date: | 2009-04-23 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct contacts between conserved motifs of different subunits provide major contribution to active site organization in human and mycobacterial dUTPases.
Febs Lett., 584, 2010
|
|
3GW6
 
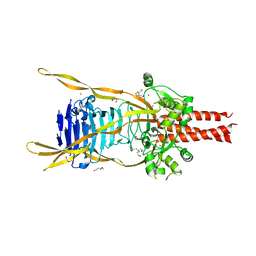 | | Intramolecular Chaperone | | Descriptor: | BROMIDE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schulz, E.C, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-03-31 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of an intramolecular chaperone mediating triple-beta-helix folding.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3H6T
 
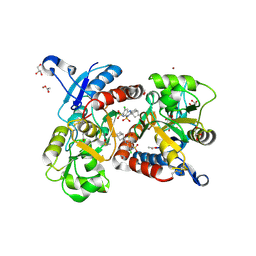 | | Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and cyclothiazide at 2.25 A resolution | | Descriptor: | ACETATE ION, CACODYLATE ION, CYCLOTHIAZIDE, ... | | Authors: | Hald, H, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2009-04-24 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Distinct structural features of cyclothiazide are responsible for effects on peak current amplitude and desensitization kinetics at iGluR2.
J.Mol.Biol., 391, 2009
|
|
3GXN
 
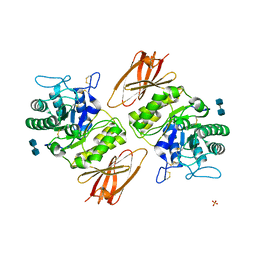 | | Crystal structure of apo alpha-galactosidase A at pH 4.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-galactosidase A, SULFATE ION | | Authors: | Lieberman, R.L. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Effects of pH and iminosugar pharmacological chaperones on lysosomal glycosidase structure and stability.
Biochemistry, 48, 2009
|
|
3H9G
 
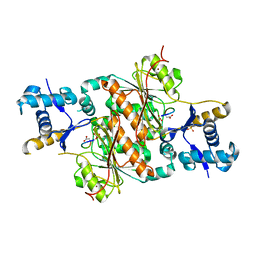 | | Crystal structure of E. coli MccB + MccA-N7isoASN | | Descriptor: | MccB protein, Microcin C7 analog, SULFATE ION, ... | | Authors: | Regni, C.A, Roush, R.F, Miller, D, Nourse, A, Walsh, C.T, Schulman, B.A. | | Deposit date: | 2009-04-30 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic.
Embo J., 28, 2009
|
|
3GYH
 
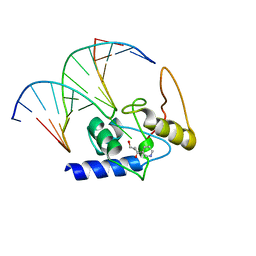 | | Crystal Structure Analysis of S. Pombe ATL in complex with damaged DNA containing POB | | Descriptor: | 1-PYRIDIN-3-YLBUTAN-1-ONE, Alkyltransferase-like protein 1, DNA (5'-D(*CP*TP*AP*CP*TP*AP*GP*CP*CP*AP*TP*GP*G)-3'), ... | | Authors: | Tubbs, J.L, Arvai, A.S, Tainer, J.A, Shin, D.S. | | Deposit date: | 2009-04-03 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Flipping of alkylated DNA damage bridges base and nucleotide excision repair.
Nature, 459, 2009
|
|
3GYO
 
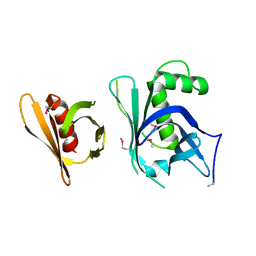 | | Se-Met Rtt106p | | Descriptor: | Histone chaperone RTT106 | | Authors: | Liu, Y, Huang, H, Shi, Y, Teng, M. | | Deposit date: | 2009-04-04 | | Release date: | 2009-12-08 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural analysis of Rtt106p reveals a DNA-binding role required for heterochromatin silencing
J.Biol.Chem., 285, 2010
|
|
3HBZ
 
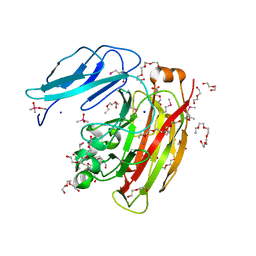 | |
3H0A
 
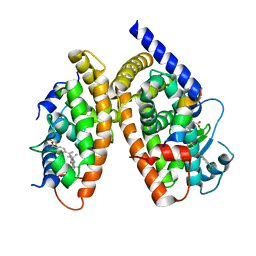 | | Crystal Structure of Peroxisome Proliferator-Activated Receptor Gamma (PPARg) and Retinoic Acid Receptor Alpha (RXRa) in Complex with 9-cis Retinoic Acid, Co-activator Peptide, and a Partial Agonist | | Descriptor: | 4-[1-(3,5,5,8,8-pentamethyl-5,6,7,8-tetrahydronaphthalen-2-yl)ethenyl]benzoic acid, Nuclear receptor coactivator 1, Co-activator Peptide, ... | | Authors: | Wang, Z, Sudom, A, Walker, N.P. | | Deposit date: | 2009-04-08 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of a PPARdelta agonist with partial agonistic activity on PPARgamma.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3K5B
 
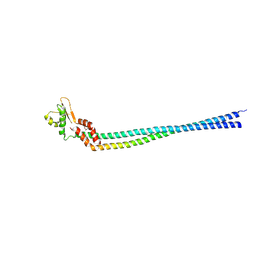 | | Crystal structure of the peripheral stalk of Thermus thermophilus H+-ATPase/synthase | | Descriptor: | V-type ATP synthase subunit E, V-type ATP synthase, subunit (VAPC-THERM) | | Authors: | Lee, L.K, Stewart, A.G, Donohoe, M, Bernal, R.A, Stock, D. | | Deposit date: | 2009-10-07 | | Release date: | 2010-02-23 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structure of the peripheral stalk of Thermus thermophilus H(+)-ATPase/synthase.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3K88
 
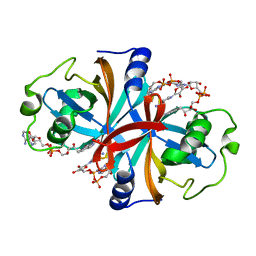 | | Crystal structure of NADH:FAD oxidoreductase (TftC) - FAD, NADH complex | | Descriptor: | Chlorophenol-4-monooxygenase component 1, FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kang, C, Webb, B.N. | | Deposit date: | 2009-10-13 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of chlorophenol 4-monooxygenase (TftD) and NADH:FAD oxidoreductase (TftC) of Burkholderia cepacia AC1100.
J.Biol.Chem., 285, 2010
|
|
3K8X
 
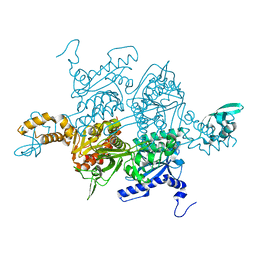 | | Crystal structure of the carboxyltransferase domain of acetyl-coenzyme A carboxylase in complex with tepraloxydim | | Descriptor: | (5S)-2-[(1E)-N-{[(2E)-3-chloroprop-2-en-1-yl]oxy}propanimidoyl]-3-hydroxy-5-(tetrahydro-2H-pyran-4-yl)cyclohex-2-en-1-one, Acetyl-CoA carboxylase | | Authors: | Xiang, S, Callaghan, M.M, Watson, K.G, Tong, L. | | Deposit date: | 2009-10-15 | | Release date: | 2009-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A different mechanism for the inhibition of the carboxyltransferase domain of acetyl-coenzyme A carboxylase by tepraloxydim.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3K9Q
 
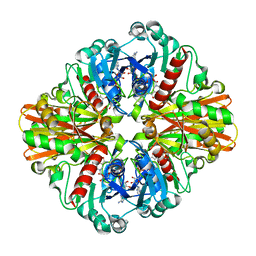 | | Crystal structure of C151G mutant of Glyceraldehyde 3-phosphate dehydrogenase 1 from Methicillin resistant Staphylococcus aureus (MRSA252) at 2.5 angstrom resolution | | Descriptor: | CHLORIDE ION, GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase 1, ... | | Authors: | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | Deposit date: | 2009-10-16 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
