3LBX
 
 | | Crystal Structure of the Erythrocyte Spectrin Tetramerization Domain Complex | | Descriptor: | Spectrin alpha chain, erythrocyte, Spectrin beta chain | | Authors: | Ipsaro, J.J, Harper, S.L, Messick, T.E, Marmorstein, R, Mondragon, A, Speicher, D.W. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure and functional interpretation of the erythrocyte spectrin tetramerization domain complex.
Blood, 115, 2010
|
|
3LD5
 
 | | Human aldose reductase mutant T113S complexed with IDD594 | | Descriptor: | Aldose reductase, BROMIDE ION, CITRIC ACID, ... | | Authors: | Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2010-01-12 | | Release date: | 2010-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Tracing the detail: how mutations affect binding modes and thermodynamic signatures of closely related aldose reductase inhibitors
J.Mol.Biol., 406, 2011
|
|
3LQV
 
 | | Branch Recognition by SF3b14 | | Descriptor: | ADENINE, Pre-mRNA branch site protein p14, Splicing factor 3B subunit 1 | | Authors: | Schellenberg, M.J, MacMillan, A.M. | | Deposit date: | 2010-02-10 | | Release date: | 2011-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Branch Recognition by SF3b14
Rna, 17, 2011
|
|
3LDH
 
 | | A comparison of the structures of apo dogfish m4 lactate dehydrogenase and its ternary complexes | | Descriptor: | LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PYRUVIC ACID | | Authors: | White, J.L, Hackert, M.L, Buehner, M, Adams, M.J, Ford, G.C, Lentzjunior, P.J, Smiley, I.E, Steindel, S.J, Rossmann, M.G. | | Deposit date: | 1974-06-06 | | Release date: | 1977-04-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A comparison of the structures of apo dogfish M4 lactate dehydrogenase and its ternary complexes.
J.Mol.Biol., 102, 1976
|
|
8ZC2
 
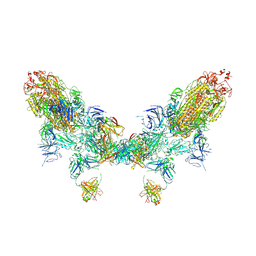 | | SARS-CoV-2 Omicron BA.2 spike trimer (6P) in complex with D1F6 Fab, head-to-head aggregate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (7.82 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZC6
 
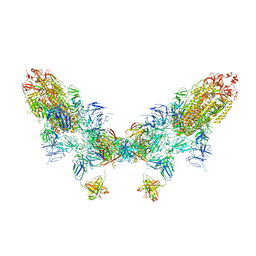 | | SARS-CoV-2 Omicron BA.4 spike trimer (6P) in complex with D1F6 Fab, head-to-head aggregate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, Light chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (6.85 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
3LSC
 
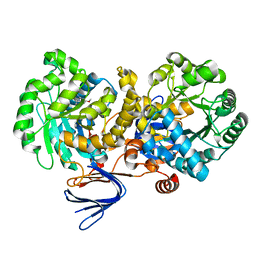 | | Crystal structure of the mutant E241Q of atrazine chlorohydrolase TrzN from Arthrobacter aurescens TC1 complexed with zinc and atraton | | Descriptor: | N-ethyl-6-methoxy-N'-(1-methylethyl)-1,3,5-triazine-2,4-diamine, Triazine hydrolase, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Seffernick, J, Wackett, L.P, Almo, S.C. | | Deposit date: | 2010-02-12 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of the mutant E241Q of atrazine chlorohydrolase
TrzN from Arthrobacter aurescens TC1 complexed with zinc and atraton
To be Published
|
|
8Z5J
 
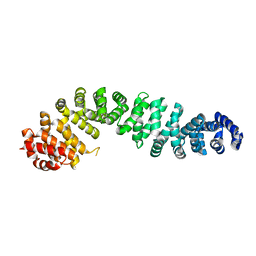 | | Beta-catenin Crystal Structure | | Descriptor: | Catenin beta-1 | | Authors: | Tim, F. | | Deposit date: | 2024-04-18 | | Release date: | 2024-05-22 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Novel 1-Phenylpiperidine Urea-Containing Derivatives Inhibiting beta-Catenin/BCL9 Interaction and Exerting Antitumor Efficacy through the Activation of Antigen Presentation of cDC1 Cells.
J.Med.Chem., 67, 2024
|
|
3T3A
 
 | | Crystal structure of H107R mutant of extracellular domain of mouse receptor NKR-P1A | | Descriptor: | Killer cell lectin-like receptor subfamily B member 1A, PHOSPHATE ION | | Authors: | Kolenko, P, Rozbesky, D, Bezouska, K, Hasek, J, Dohnalek, J. | | Deposit date: | 2011-07-25 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the H107R variant of the extracellular domain of mouse NKR-P1A at 2.3 A resolution.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3LGN
 
 | | Crystal structure of IsdI in complex with heme | | Descriptor: | Heme-degrading monooxygenase isdI, MAGNESIUM ION, OXYGEN MOLECULE, ... | | Authors: | Ukpabi, G.N, Murphy, M.E.P. | | Deposit date: | 2010-01-20 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The IsdG-family of haem oxygenases degrades haem to a novel chromophore
Mol.Microbiol., 75, 2010
|
|
8JC4
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A-T268V in complex with Pyd-Pid-Phe and hydroxylamine | | Descriptor: | 1-pyridin-4-ylpiperidine-4-carboxylic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2023-05-10 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | C(sp3)-H hydroxylation of Broad-Spectrum Alkanes Catalyzed by an Artificial P450 Peroxygenase Driven by omega-Pyridyl Fatty Acyl Amino Acids.
Mol Catal, 550, 2023
|
|
8JC3
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A-T268V in complex with Pyd-N-C4-Phe and hydroxylamine | | Descriptor: | 4-(pyridin-4-ylamino)butanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2023-05-10 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | C(sp3)-H hydroxylation of Broad-Spectrum Alkanes Catalyzed by an Artificial P450 Peroxygenase Driven by omega-Pyridyl Fatty Acyl Amino Acids.
Mol Catal, 550, 2023
|
|
3LPJ
 
 | | Structure of BACE Bound to SCH743641 | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N'-[(1S,2S)-2-[(2R)-4-benzylpiperazin-2-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2010-02-05 | | Release date: | 2010-04-14 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3LQ5
 
 | | Structure of CDK9/CyclinT in complex with S-CR8 | | Descriptor: | (2S)-2-({9-(1-methylethyl)-6-[(4-pyridin-2-ylbenzyl)amino]-9H-purin-2-yl}amino)butan-1-ol, Cell division protein kinase 9, Cyclin-T1 | | Authors: | Hole, A.J, Endicott, J.A, Baumli, S. | | Deposit date: | 2010-02-08 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | CDK Inhibitors Roscovitine and CR8 Trigger Mcl-1 Down-Regulation and Apoptotic Cell Death in Neuroblastoma Cells
Genes Cancer, 1, 2010
|
|
7BR7
 
 | | Epstein-Barr virus, C1 portal-proximal penton vertex, CATC binding | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | CryoEM structure of the tegumented capsid of Epstein-Barr virus.
Cell Res., 30, 2020
|
|
8D3D
 
 | | VWF tubule derived from dimeric D1-A1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand factor | | Authors: | Anderson, J.R, Li, J, Springer, T.A, Brown, A. | | Deposit date: | 2022-06-01 | | Release date: | 2022-07-06 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of VWF tubules before and after concatemerization reveal a mechanism of disulfide bond exchange.
Blood, 140, 2022
|
|
3LS9
 
 | | Crystal structure of atrazine chlorohydrolase TrzN from Arthrobacter aurescens TC1 complexed with zinc | | Descriptor: | Triazine hydrolase, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Seffernick, J, Wackett, L.P, Almo, S.C. | | Deposit date: | 2010-02-12 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of atrazine chlorohydrolase TrzN
from Arthrobacter aurescens TC1 complexed with zinc
To be Published
|
|
3LY1
 
 | |
3T25
 
 | | TMAO-grown orthorhombic trypsin (bovine) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Cahn, J, Venkat, M, Marshall, H, Juers, D. | | Deposit date: | 2011-07-22 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The use of trimethylamine N-oxide as a primary precipitating agent and related methylamine osmolytes as cryoprotective agents for macromolecular crystallography.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3LZ3
 
 | | Human aldose reductase mutant T113S complexed with IDD388 | | Descriptor: | (2-{[(4-BROMO-2-FLUOROBENZYL)AMINO]CARBONYL}-5-CHLOROPHENOXY)ACETIC ACID, Aldose reductase, BROMIDE ION, ... | | Authors: | Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2010-03-01 | | Release date: | 2010-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Tracing the detail: how mutations affect binding modes and thermodynamic signatures of closely related aldose reductase inhibitors
J.Mol.Biol., 406, 2011
|
|
3L62
 
 | | Crystal structure of substrate-free P450cam at low [K+] | | Descriptor: | Camphor 5-monooxygenase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lee, Y.-T, Wilson, R.F, Rupniewski, I, Goodin, D.B. | | Deposit date: | 2009-12-22 | | Release date: | 2010-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | P450cam visits an open conformation in the absence of substrate.
Biochemistry, 49, 2010
|
|
8XJ3
 
 | |
8D6W
 
 | |
8D6V
 
 | |
8ZNC
 
 | |
