3IHB
 
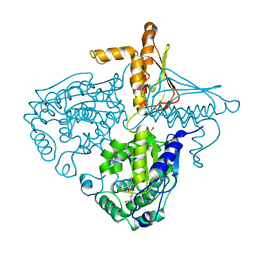 | | Crystal Structure Analysis of Mglu in its tris and glutamate form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLUTAMIC ACID, Salt-tolerant glutaminase | | Authors: | Yoshimune, K, Shirakihara, Y. | | Deposit date: | 2009-07-29 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of salt-tolerant glutaminase from Micrococcus luteus K-3 in the presence and absence of its product l-glutamate and its activator Tris
Febs J., 277, 2010
|
|
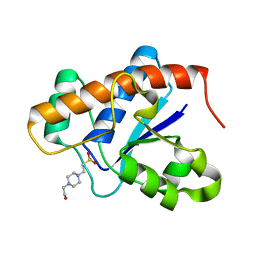
3IDO
 
 | |
3IDZ
 
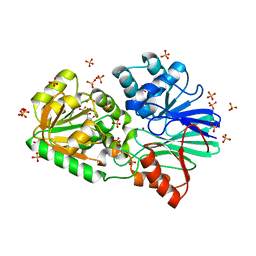 | | Crystal Structure of S378Q mutant TTHA0252 from Thermus thermophilus HB8 | | Descriptor: | CITRATE ANION, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of S378Q mutant TTHA0252 from Thermus thermophilus HB8
To be Published
|
|
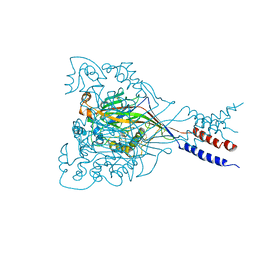
3IJ4
 
 | |
3IER
 
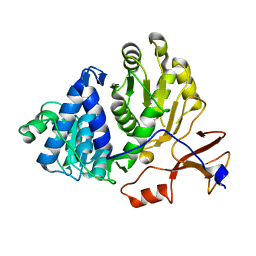 | | Firefly luciferase apo structure (P41 form) with PEG 400 bound | | Descriptor: | Luciferin 4-monooxygenase, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Battaile, K.P, Auld, D.S, Thorne, N, Lea, W.A, Maloney, D.J, Shen, M, Raj, G, Thomas, C.J, Simeonov, A, Hanzlik, R.P, Inglese, J. | | Deposit date: | 2009-07-23 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular basis for the high-affinity binding and stabilization of firefly luciferase by PTC124.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3IF5
 
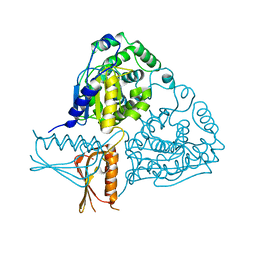 | | Crystal Structure Analysis of Mglu | | Descriptor: | Salt-tolerant glutaminase | | Authors: | Yoshimune, K, Shirakihara, Y. | | Deposit date: | 2009-07-24 | | Release date: | 2009-08-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of salt-tolerant glutaminase from Micrococcus luteus K-3 in the presence and absence of its product L-glutamate and its activator Tris.
Febs J., 277, 2010
|
|
3IJG
 
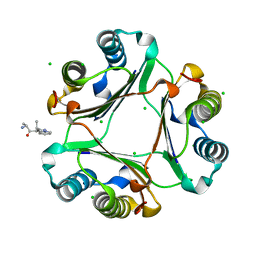 | | Macrophage Migration Inhibitory Factor (MIF) Bound to the (R)-Stereoisomer of AV1013 | | Descriptor: | (2R)-2-amino-1-[2-(1-methylethyl)pyrazolo[1,5-a]pyridin-3-yl]propan-1-one, CHLORIDE ION, Macrophage migration inhibitory factor | | Authors: | Crichlow, G.V, Cho, Y, Lolis, E.J. | | Deposit date: | 2009-08-04 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Allosteric inhibition of macrophage migration inhibitory factor revealed by ibudilast.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3IFJ
 
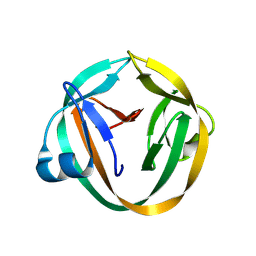 | |
3IKE
 
 | |
3IG7
 
 | | Novel CDK-5 inhibitors - crystal structure of inhibitor EFP with CDK-2 | | Descriptor: | Cell division protein kinase 2, N-{1-[cis-3-(acetylamino)cyclobutyl]-1H-imidazol-4-yl}-2-(4-methoxyphenyl)acetamide | | Authors: | Pandit, J. | | Deposit date: | 2009-07-27 | | Release date: | 2009-09-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent and cellularly active 4-aminoimidazole inhibitors of cyclin-dependent kinase 5/p25 for the treatment of Alzheimer's disease.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3IMM
 
 | |
3IH7
 
 | |
3INO
 
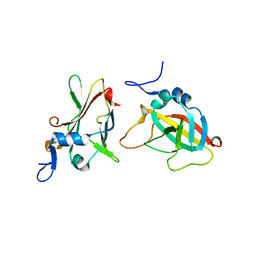 | | 1.95A Resolution Structure of Protective Antigen Domain 4 | | Descriptor: | Protective antigen PA-63 | | Authors: | Lovell, S, Williams, A.S, Anbanandam, A, El-Chami, R, Bann, J.G. | | Deposit date: | 2009-08-12 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Domain 4 of the anthrax protective antigen maintains structure and binding to the host receptor CMG2 at low pH
Protein Sci., 18, 2009
|
|
3II4
 
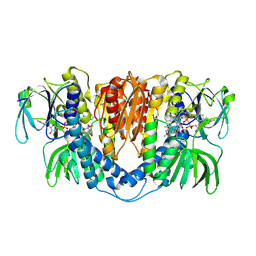 | |
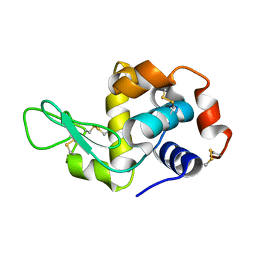
3IJV
 
 | |
3IJX
 
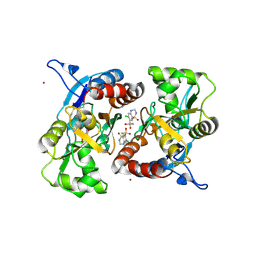 | | Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, hydrochlorothiazide | | Descriptor: | 6-chloro-3,4-dihydro-2H-1,2,4-benzothiadiazine-7-sulfonamide 1,1-dioxide, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Ptak, C.P, Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2009-08-05 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.881 Å) | | Cite: | Probing the allosteric modulator binding site of GluR2 with thiazide derivatives
Biochemistry, 48, 2009
|
|
3IKF
 
 | | Crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase from Burkholderia pseudomallei with FOL fragment 717, imidazo[2,,1-b][1,3]thiazol-6-ylmethanol | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-08-05 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Leveraging structure determination with fragment screening for infectious disease drug targets: MECP synthase from Burkholderia pseudomallei.
J Struct Funct Genomics, 12, 2011
|
|
3IPH
 
 | | Crystal structure of p38 in complex with a biphenylamide inhibitor | | Descriptor: | 6-[5-(cyclopropylcarbamoyl)-2-methylphenyl]-N-(cyclopropylmethyl)pyridine-3-carboxamide, GLYCEROL, Mitogen-activated protein kinase 14, ... | | Authors: | Somers, D.O. | | Deposit date: | 2009-08-17 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | p38alpha mitogen-activated protein kinase inhibitors: optimization of a series of biphenylamides to give a molecule suitable for clinical progression.
J.Med.Chem., 52, 2009
|
|
3IMU
 
 | |
3IN9
 
 | | Crystal structure of heparin lyase I complexed with disaccharide heparin | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, Heparin lyase I | | Authors: | Han, Y.H, Ryu, K.S, Kim, H.Y, Jeon, Y.H. | | Deposit date: | 2009-08-12 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural snapshots of heparin depolymerization by heparin lyase I
J.Biol.Chem., 284, 2009
|
|
3IQQ
 
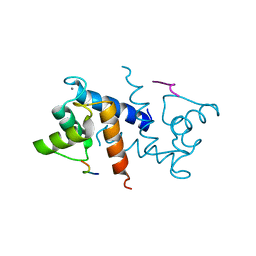 | | X-ray structure of bovine TRTK12-Ca(2+)-S100B | | Descriptor: | CALCIUM ION, Protein S100-B, TRTK12 peptide, ... | | Authors: | Charpentier, T.H, Weber, D.J, Toth, E.A. | | Deposit date: | 2009-08-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Effects of CapZ Peptide (TRTK-12) Binding to S100B-Ca(2+) as Examined by NMR and X-ray Crystallography
J.Mol.Biol., 396, 2010
|
|
3IQX
 
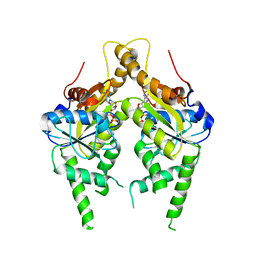 | | ADP complex of C.therm. Get3 in closed form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Tail-anchored protein targeting factor Get3, ... | | Authors: | Bozkurt, G, Wild, K, Sinning, I. | | Deposit date: | 2009-08-21 | | Release date: | 2009-12-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural insights into tail-anchored protein binding and membrane insertion by Get3.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3IRC
 
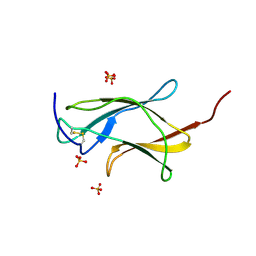 | | Crystal structure analysis of dengue-1 envelope protein domain III | | Descriptor: | ENVELOPE PROTEIN, SULFATE ION | | Authors: | Nelson, C.A, Kim, T, Warren, J.T, Chruszcz, M, Minor, W, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-21 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure Analysis of the Dengue-1 Envelope Protein Domain III
To be Published
|
|
3ISE
 
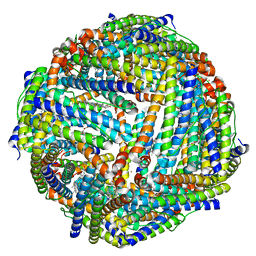 | | Structure of mineralized Bfrb (double soak) from Pseudomonas aeruginosa to 2.8A Resolution | | Descriptor: | Bacterioferritin, FE (III) ION, POTASSIUM ION, ... | | Authors: | Lovell, S, Weeratunga, S.K, Battaile, K.P, Rivera, M. | | Deposit date: | 2009-08-25 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Studies of Bacterioferritin B from Pseudomonas aeruginosa Suggest a Gating Mechanism for Iron Uptake via the Ferroxidase Center
Biochemistry, 49, 2010
|
|
3IT0
 
 | | Crystal Structure Francisella tularensis histidine acid phosphatase complexed with phosphate | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Acid phosphatase, PENTAETHYLENE GLYCOL, ... | | Authors: | Singh, H, Felts, R.L, Reilly, T.J, Tanner, J.J. | | Deposit date: | 2009-08-27 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | Crystal Structures of the histidine acid phosphatase from Francisella tularensis provide insight into substrate recognition.
J.Mol.Biol., 394, 2009
|
|
