1BW6
 
 | | HUMAN CENTROMERE PROTEIN B (CENP-B) DNA BINDIGN DOMAIN RP1 | | Descriptor: | PROTEIN (CENTROMERE PROTEIN B) | | Authors: | Iwahara, J, Kigawa, T, Kitagawa, K, Masumoto, H, Okazaki, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-09-30 | | Release date: | 1998-10-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A helix-turn-helix structure unit in human centromere protein B (CENP-B).
EMBO J., 17, 1998
|
|
1QNU
 
 | | Shiga-Like Toxin I B Subunit Complexed with the Bridged-Starfish Inhibitor | | Descriptor: | ETHYL-CARBAMIC ACID METHYL ESTER, METHYL-CARBAMIC ACID ETHYL ESTER, Shiga toxin 1 variant B subunit, ... | | Authors: | Pannu, N.S, Hayakawa, K, Read, R.J. | | Deposit date: | 1999-10-21 | | Release date: | 2000-04-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Shiga-like toxins are neutralized by tailored multivalent carbohydrate ligands.
Nature, 403, 2000
|
|
2X7G
 
 | | Structure of human serine-arginine-rich protein-specific kinase 2 (SRPK2) bound to purvalanol B | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, PURVALANOL B, ... | | Authors: | Pike, A.C.W, Savitsky, P, Fedorov, O, Krojer, T, Ugochukwu, E, von Delft, F, Gileadi, O, Edwards, A, Arrowsmith, C.H, Weigelt, J, Bountra, C, Knapp, S. | | Deposit date: | 2010-02-26 | | Release date: | 2010-04-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Human Serine-Arginine-Rich Protein- Specific Kinase 2 (Srpk2) Bound to Purvalanol B
To be Published
|
|
2NM1
 
 | | Structure of BoNT/B in complex with its protein receptor | | Descriptor: | Botulinum neurotoxin type B, Synaptotagmin-2 | | Authors: | Jin, R, Rummel, A, Binz, T, Brunger, A.T. | | Deposit date: | 2006-10-20 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Botulinum neurotoxin B recognizes its protein receptor with high affinity and specificity.
Nature, 444, 2006
|
|
7CMK
 
 | | E30 E-particle in complex with 6C5 | | Descriptor: | Heavy chain, Light chain, VP1, ... | | Authors: | Wang, K, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-07-27 | | Release date: | 2020-08-12 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Serotype specific epitopes identified by neutralizing antibodies underpin immunogenic differences in Enterovirus B.
Nat Commun, 11, 2020
|
|
6KH8
 
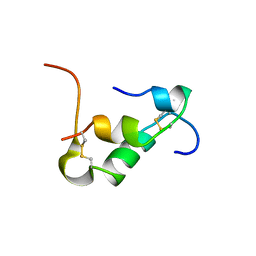 | | Solution structure of Zn free Bovine Pancreatic Insulin in 20% acetic acid-d4 (pH 1.9) | | Descriptor: | Insulin A Chain, Insulin B chain | | Authors: | Bhunia, A, Ratha, B.N, Kar, R.K, Brender, J.R. | | Deposit date: | 2019-07-14 | | Release date: | 2020-10-07 | | Last modified: | 2020-11-18 | | Method: | SOLUTION NMR | | Cite: | High-resolution structure of a partially folded insulin aggregation intermediate.
Proteins, 88, 2020
|
|
1OGB
 
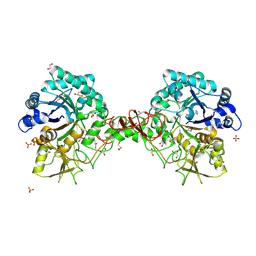 | | Chitinase b from Serratia marcescens mutant D142N | | Descriptor: | CHITINASE B, GLYCEROL, SULFATE ION | | Authors: | Vaaje-Kolstad, G, Houston, D.R, Rao, F.V, Peter, M.G, Synstad, B, van Aalten, D.M.F, Eijsink, V.G.H. | | Deposit date: | 2003-04-29 | | Release date: | 2004-04-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the D142N Mutant of the Family 18 Chitinase Chib from Serratia Marcescens and its Complex with Allosamidin
Biochim.Biophys.Acta, 1696, 2004
|
|
6KH9
 
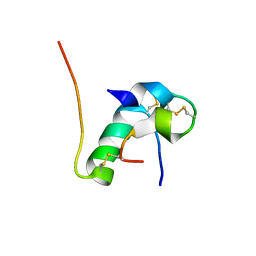 | | Solution structure of bovine insulin amyloid intermediate-1 | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Ratha, B.N, Kar, R.K, Brender, J.B, Bhunia, A. | | Deposit date: | 2019-07-14 | | Release date: | 2020-08-12 | | Last modified: | 2020-11-18 | | Method: | SOLUTION NMR | | Cite: | High-resolution structure of a partially folded insulin aggregation intermediate.
Proteins, 88, 2020
|
|
4YTF
 
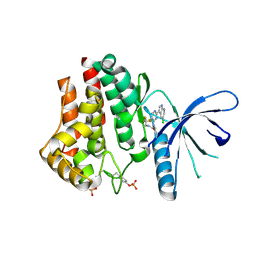 | | Discovery of VX-509 (Decernotinib): A Potent and Selective Janus kinase (JAK) 3 Inhibitor for the Treatment of Autoimmune Diseases | | Descriptor: | N~2~-[2-(5-chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)-5-fluoropyrimidin-4-yl]-N-(2,2,2-trifluoroethyl)-L-alaninamide, Tyrosine-protein kinase JAK2 | | Authors: | Farmer, L, Ledeboer, M.W, Zuccola, H.J. | | Deposit date: | 2015-03-17 | | Release date: | 2015-08-12 | | Last modified: | 2015-10-07 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Discovery of VX-509 (Decernotinib): A Potent and Selective Janus Kinase 3 Inhibitor for the Treatment of Autoimmune Diseases.
J.Med.Chem., 58, 2015
|
|
1UZ0
 
 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with Glc-4Glc-3Glc-4Glc | | Descriptor: | CALCIUM ION, CELLULASE B, CHLORIDE ION, ... | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
4YTI
 
 | |
1UYY
 
 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with cellotriose | | Descriptor: | CALCIUM ION, CELLULASE B, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
4MAX
 
 | | Crystal structure of Synechococcus sp. PCC 7002 globin at cryogenic temperature with heme modification | | Descriptor: | HEME B/C, SULFATE ION, cyanoglobin | | Authors: | Wenke, B.B, Schlessman, J.L, Heroux, A, Lecomte, J.T.J. | | Deposit date: | 2013-08-18 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.444 Å) | | Cite: | The 2/2 hemoglobin from the cyanobacterium Synechococcus sp. PCC 7002 with covalently attached heme: Comparison of X-ray and NMR structures.
Proteins, 82, 2014
|
|
7YC9
 
 | | Co-crystal structure of BTK kinase domain with inhibitor | | Descriptor: | (7~{S})-2-(4-bromanyl-3,5-dimethoxy-phenyl)-7-(1-propanoylpiperidin-4-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, 1,2-ETHANEDIOL, Tyrosine-protein kinase BTK | | Authors: | Zhou, X. | | Deposit date: | 2022-07-01 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of BGB-8035, a Highly Selective Covalent Inhibitor of Bruton's Tyrosine Kinase for B-Cell Malignancies and Autoimmune Diseases.
J.Med.Chem., 66, 2023
|
|
7G19
 
 | | Crystal Structure of human FABP4 in complex with 5-[2-(1H-tetrazol-5-yl)ethyl]-6,7,8,9,10,11-hexahydrocycloocta[b]indole | | Descriptor: | 5-[2-(1H-tetrazol-5-yl)ethyl]-6,7,8,9,10,11-hexahydro-5H-cycloocta[b]indole, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Kyburz, E, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
1LEC
 
 | | STRUCTURES OF THE LECTIN IV OF GRIFFONIA SIMPLICIFOLIA AND ITS COMPLEX WITH THE LEWIS B HUMAN BLOOD GROUP DETERMINANT AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Delbaere, L, Vandonselaar, M, Quail, J. | | Deposit date: | 1992-12-17 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the lectin IV of Griffonia simplicifolia and its complex with the Lewis b human blood group determinant at 2.0 A resolution.
J.Mol.Biol., 230, 1993
|
|
7CWA
 
 | | Crystal structure of PDE8A catalytic domain in complex with clofarabine | | Descriptor: | 2-CHLORO-9-(2-DEOXY-2-FLUORO-B -D-ARABINOFURANOSYL)-9H-PURIN-6-AMINE, High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A, MAGNESIUM ION, ... | | Authors: | Huang, Y, Wu, X.-N, Zhou, Q, Wu, Y, Luo, H.-B. | | Deposit date: | 2020-08-27 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Rational Design of 2-Chloroadenine Derivatives as Highly Selective Phosphodiesterase 8A Inhibitors.
J.Med.Chem., 63, 2020
|
|
7GZG
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000620-001 | | Descriptor: | (3R)-3-(4-bromophenyl)-3-[(1H-pyrrolo[3,2-b]pyridine-5-carbonyl)amino]propanoic acid, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7GZF
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000605-001 | | Descriptor: | (3R)-3-(4-bromophenyl)-3-[(1-methyl-1H-pyrazolo[3,4-b]pyridine-4-carbonyl)amino]propanoic acid, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7GZI
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000753-001 | | Descriptor: | 3-[4-(cyclopropylcarbamamido)benzamido]-1-methyl-1H-pyrrolo[2,3-b]pyridine-2-carboxylic acid, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7H0B
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0010716-001 | | Descriptor: | (4M)-4-(4-{[(1R)-1-(2,3-dihydro[1,4]dioxino[2,3-b]pyridin-6-yl)-2-methylpropyl]amino}-7H-pyrrolo[2,3-d]pyrimidin-6-yl)-1-methyl-1H-pyrazole-5-carbonitrile, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
6B3T
 
 | | Crystal structure of acetyltransferase Eis from Mycobacterium tuberculosis in complex with a 1,2,4-triazino[5,6b]indole-3-thioether inhibitor analogue 39b | | Descriptor: | 8-fluoro-5-methyl-3-{[2-(piperidin-1-yl)ethyl]sulfanyl}-5H-[1,2,4]triazino[5,6-b]indole, COENZYME A, N-acetyltransferase Eis, ... | | Authors: | Gajadeera, C.S, Hou, C, Garneau-Tsodikova, S, Ngo, H.X, Tsodikov, O.V. | | Deposit date: | 2017-09-24 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Potent 1,2,4-Triazino[5,6 b]indole-3-thioether Inhibitors of the Kanamycin Resistance Enzyme Eis from Mycobacterium tuberculosis.
ACS Infect Dis, 4, 2018
|
|
7GZH
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000670-001 | | Descriptor: | (3R)-3-(4-bromophenyl)-3-[(1H-pyrrolo[2,3-b]pyridine-5-carbonyl)amino]propanoic acid, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
5EZV
 
 | |
7YC2
 
 | |
