5EA6
 
 | | Crystal Structure of Inhibitor BTA-9881 in Complex with Prefusion RSV F Glycoprotein | | Descriptor: | (9~{b}~{R})-9~{b}-(4-chlorophenyl)-1-pyridin-3-ylcarbonyl-2,3-dihydroimidazo[5,6]pyrrolo[1,2-~{a}]pyridin-5-one, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Fusion glycoprotein F0, ... | | Authors: | Battles, M.B, McLellan, J.S, Arnoult, E, Roymans, D, Langedijk, J.P. | | Deposit date: | 2015-10-15 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Molecular mechanism of respiratory syncytial virus fusion inhibitors.
Nat.Chem.Biol., 12, 2016
|
|
4NRK
 
 | | Structure of hemagglutinin with F95Y mutation of influenza virus B/Lee/40 complex with LSTc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Ni, F, Mbawuike, I.N, Kondrashkina, E, Wang, Q. | | Deposit date: | 2013-11-26 | | Release date: | 2014-03-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | The roles of hemagglutinin Phe-95 in receptor binding and pathogenicity of influenza B virus.
Virology, 450-451, 2014
|
|
5FZ9
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with Maybridge fragment thieno(3,2-b)thiophene-5-carboxylic acid (N06263b) (ligand modelled based on PANDDA event map, SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Kupinska, K, Szykowska, A, Pearce, N, Talon, R, Collins, P, Gileadi, C, Strain-Damerell, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-12 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with Maybridge Fragment Thieno(3,2-B)Thiophene -5-Carboxylic Acid (N06263B) (Ligand Modelled Based on Pandda Event Map, Sgc - Diamond I04-1 Fragment Screening)
To be Published
|
|
7BPI
 
 | | The crystal structue of PDE10A complexed with 14 | | Descriptor: | 8-[(E)-2-[5-methyl-1-[3-[3-(4-methylpiperazin-1-yl)propoxy]phenyl]benzimidazol-2-yl]ethenyl]quinoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Yang, Y, Zhang, S, Zhou, Q, Huang, Y.-Y, Guo, L, Luo, H.-B. | | Deposit date: | 2020-03-22 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4000864 Å) | | Cite: | Discovery of highly selective and orally available benzimidazole-based phosphodiesterase 10 inhibitors with improved solubility and pharmacokinetic properties for treatment of pulmonary arterial hypertension.
Acta Pharm Sin B, 10, 2020
|
|
1UY0
 
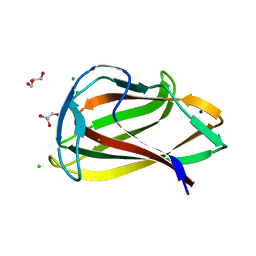 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with glc-1,3-glc-1,4-glc-1,3-glc | | Descriptor: | CALCIUM ION, CELLULASE B, CHLORIDE ION, ... | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-01 | | Release date: | 2004-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
1RK1
 
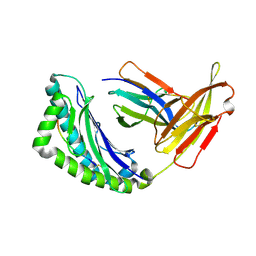 | | Mhc Class I Natural H-2Kb Heavy Chain Complexed With beta-2 Microglobulin and Herpes Simplex Virus Mutant Glycoprotein B Peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Glycoprotein B, ... | | Authors: | Miley, M.J, Messaoudi, I, Nikolich-Zugich, J, Fremont, D.H. | | Deposit date: | 2003-11-20 | | Release date: | 2004-12-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for the Restoration of TCR Recognition of an MHC Allelic Variant by Peptide Secondary Anchor Substitution
J.Exp.Med., 200, 2004
|
|
4MS1
 
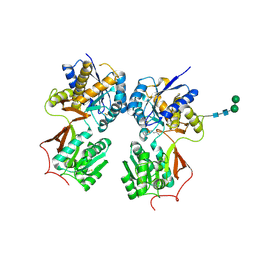 | | Crystal structure of the extracellular domain of human GABA(B) receptor bound to the antagonist CGP46381 | | Descriptor: | (S)-(3-aminopropyl)(cyclohexylmethyl)phosphinic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Geng, Y, Bush, M, Mosyak, L, Wang, F, Fan, Q.R. | | Deposit date: | 2013-09-18 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural mechanism of ligand activation in human GABA(B) receptor.
Nature, 504, 2013
|
|
2XCQ
 
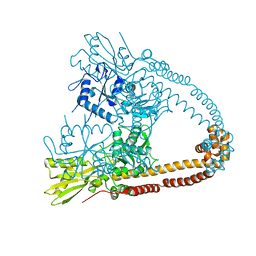 | | The 2.98A crystal structure of the catalytic core (B'A' region) of Staphylococcus aureus DNA Gyrase | | Descriptor: | DNA GYRASE SUBUNIT B, DNA GYRASE SUBUNIT A | | Authors: | Bax, B.D, Chan, P.F, Eggleston, D.S, Fosberry, A, Gentry, D.R, Gorrec, F, Giordano, I, Hann, M.M, Hennessy, A, Hibbs, M, Huang, J, Jones, E, Jones, J, Brown, K.K, Lewis, C.J, May, E.W, Singh, O, Spitzfaden, C, Shen, C, Shillings, A, Theobald, A.F, Wohlkonig, A, Pearson, N.D, Gwynn, M.N. | | Deposit date: | 2010-04-24 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Type Iia Topoisomerase Inhibition by a New Class of Antibacterial Agents.
Nature, 466, 2010
|
|
6BZ2
 
 | | Crystal structure of wild-type HIV-1 protease with a novel HIV-1 inhibitor GRL-14213A of 6-5-5-ring fused crown-like tetrahydropyranofuran as the P2-ligand, a cyclopropylaminobenzothiazole as the P2'-ligand and 3,5-difluorophenylmethyl as the P1-ligand | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2017-12-22 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Design of Highly Potent, Dual-Acting and Central-Nervous-System-Penetrating HIV-1 Protease Inhibitors with Excellent Potency against Multidrug-Resistant HIV-1 Variants.
ChemMedChem, 13, 2018
|
|
4KBD
 
 | | DNA STRUCTURE OF A MUTATED KB SITE | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*GP*AP*AP*AP*GP*TP*GP*AP*GP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*CP*TP*CP*AP*CP*TP*TP*TP*CP*CP*AP*GP*G)-3') | | Authors: | Tisne, C, Hartmann, B, Delepierre, M. | | Deposit date: | 1998-11-30 | | Release date: | 1999-10-14 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NF-kappa B binding mechanism: a nuclear magnetic resonance and modeling study of a GGG --> CTC mutation.
Biochemistry, 38, 1999
|
|
5TKD
 
 | | CRYSTAL STRUCTURE OF TYROSINE KINASE 2 JH2 (PSEUDO KINASE DOMAIN) COMPLEXED WITH 6-[(3,5-DIMETHYLPHE NYL)AMINO]-8- (METHYLAMINO)IMIDAZO[1,2-B]PYRIDAZINE-3-CARBO XAMIDE | | Descriptor: | 6-[(3,5-dimethylphenyl)amino]-8-(methylamino)imidazo[1,2-b]pyridazine-3-carboxamide, Non-receptor tyrosine-protein kinase TYK2, SULFATE ION | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2016-10-06 | | Release date: | 2016-12-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Identification of imidazo[1,2-b]pyridazine TYK2 pseudokinase ligands as potent and selective allosteric inhibitors of TYK2 signalling.
Medchemcomm, 8, 2017
|
|
1M6O
 
 | | Crystal Structure of HLA B*4402 in complex with HLA DPA*0201 peptide | | Descriptor: | Beta-2-microglobulin, HLA DPA*0201 peptide, HLA class I histocompatibility antigen, ... | | Authors: | Macdonald, W.A, Purcell, A.W, Williams, D.S, Mifsud, N.A, Ely, L.K, Gorman, J.J, Clements, C.S, Kjer-Nielsen, L, Koelle, D.M, Brooks, A.G, Lovrecz, G.O, Lu, L, Rossjohn, J, McCluskey, J. | | Deposit date: | 2002-07-17 | | Release date: | 2003-09-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A naturally selected dimorphism within the HLA-B44 supertype alters class I structure, peptide repertoire, and T cell recognition.
J.Exp.Med., 198, 2003
|
|
1CTE
 
 | |
4M44
 
 | | Crystal structure of hemagglutinin of influenza virus B/Yamanashi/166/1998 in complex with avian-like receptor LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Ni, F, Kondrashkina, E, Wang, Q. | | Deposit date: | 2013-08-06 | | Release date: | 2013-09-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the divergent evolution of influenza B virus hemagglutinin.
Virology, 446, 2013
|
|
5E9X
 
 | |
1RK0
 
 | | Mhc Class I H-2Kb Heavy Chain Complexed With beta-2 Microglobulin and Herpes Simplex Virus Glycoprotein B peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Glycoprotein B, ... | | Authors: | Miley, M.J, Messaoudi, I, Nikolich-Zugich, J, Fremont, D.H. | | Deposit date: | 2003-11-20 | | Release date: | 2004-12-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural Basis for the Restoration of TCR Recognition of an MHC Allelic Variant by Peptide Secondary Anchor Substitution
J.Exp.Med., 200, 2004
|
|
5VA4
 
 | |
4R5N
 
 | | 8-Tetrahydropyran-2-yl chromans: highly selective beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) inhibitors | | Descriptor: | (4R,4a'S,10a'R)-8'-(2-fluoropyridin-3-yl)-4a'-methyl-3',4',4a',10a'-tetrahydro-2'H-spiro[1,3-oxazole-4,10'-pyrano[3,2-b]chromen]-2-amine, Beta-secretase 1, NICKEL (II) ION | | Authors: | Vigers, G.P.A, Smith, D. | | Deposit date: | 2014-08-21 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 8-Tetrahydropyran-2-yl Chromans: Highly Selective Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors.
J.Med.Chem., 57, 2014
|
|
6C7Q
 
 | | BRD4 BD2 in complex with compound CE277 | | Descriptor: | 7-(3,5-dimethyl-1,2-oxazol-4-yl)-6-methoxy-2-methyl-N-(1-methyl-1H-indazol-3-yl)-9H-pyrimido[4,5-b]indol-4-amine, Bromodomain-containing protein 4 | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2018-01-23 | | Release date: | 2018-08-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure-Based Discovery of CF53 as a Potent and Orally Bioavailable Bromodomain and Extra-Terminal (BET) Bromodomain Inhibitor.
J. Med. Chem., 61, 2018
|
|
1W0W
 
 | | Crystal Structure Of HLA-B*2709 Complexed With the self-Peptide TIS from EGF-response factor 1 | | Descriptor: | BETA-2-MICROGLOBULIN, BUTYRATE RESPONSE FACTOR 2, GLYCEROL, ... | | Authors: | Hulsmeyer, M, Fiorillo, M.T, Bettosini, F, Sorrentino, R, Saenger, W, Ziegler, A, Uchanska-Ziegler, B. | | Deposit date: | 2004-06-14 | | Release date: | 2005-03-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Thermodynamic and Structural Equivalence of Two Hla-B27 Subtypes Complexed with a Self-Peptide
J.Mol.Biol., 346, 2005
|
|
6V4T
 
 | | MPER-TMD of HIV-1 Env bound with the entry inhibitor S2C3 | | Descriptor: | 4,4'-(decane-1,10-diyl)bis(9-amino-2,3-dihydro-1H-cyclopenta[b]quinolin-4-ium), Envelope glycoprotein gp160 | | Authors: | Xiao, T, Frey, G, Fu, Q, Lavine, C.L, Scott, D.A, Seaman, M.S, Chou, J.J, Chen, B. | | Deposit date: | 2019-11-30 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | HIV-1 fusion inhibitors targeting the membrane-proximal external region of Env spikes.
Nat.Chem.Biol., 16, 2020
|
|
7ZJQ
 
 | |
1DBO
 
 | | CRYSTAL STRUCTURE OF CHONDROITINASE B | | Descriptor: | 4-deoxy-alpha-D-glucopyranose-(1-3)-[beta-D-glucopyranose-(1-4)]2-O-methyl-beta-L-fucopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-2)-[alpha-L-rhamnopyranose-(1-4)]alpha-D-mannopyranose, 4-deoxy-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, CHONDROITINASE B | | Authors: | Huang, W, Matte, A, Li, Y, Kim, Y.S, Linhardt, R.J, Su, H, Cygler, M. | | Deposit date: | 1999-11-03 | | Release date: | 2000-01-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of chondroitinase B from Flavobacterium heparinum and its complex with a disaccharide product at 1.7 A resolution.
J.Mol.Biol., 294, 1999
|
|
5TWM
 
 | | CRYSTAL STRUCTURE OF THE HEPATITIS C VIRUS GENOTYPE 2A STRAIN JFH1 L30S NS5B RNA-DEPENDENT RNA POLYMERASE IN COMPLEX WITH 5-[3-(tert-butylcarbamoyl)phenyl]-6-(ethylamino)-2-(4-fluorophenyl)-N-methylfuro[2,3-b]pyridine-3-carboxamide | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, 5-[3-(tert-butylcarbamoyl)phenyl]-6-(ethylamino)-2-(4-fluorophenyl)-N-methylfuro[2,3-b]pyridine-3-carboxamide, NS5B RNA-dependent RNA POLYMERASE, ... | | Authors: | Sheriff, S. | | Deposit date: | 2016-11-14 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The discovery of a pan-genotypic, primer grip inhibitor of HCV NS5B polymerase.
Medchemcomm, 8, 2017
|
|
6C7R
 
 | | BRD4 BD1 in complex with compound CF53 | | Descriptor: | Bromodomain-containing protein 4, N-(3-cyclopropyl-1-methyl-1H-pyrazol-5-yl)-7-(3,5-dimethyl-1,2-oxazol-4-yl)-6-methoxy-2-methyl-9H-pyrimido[4,5-b]indol-4-amine | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2018-01-23 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Discovery of CF53 as a Potent and Orally Bioavailable Bromodomain and Extra-Terminal (BET) Bromodomain Inhibitor.
J. Med. Chem., 61, 2018
|
|
