3FMD
 
 | | Crystal Structure of Human Haspin with an Isoquinoline ligand | | Descriptor: | 1,2-ETHANEDIOL, N-[2-(4-BROMOCINNAMYLAMINO)ETHYL]-5-ISOQUINOLINE SULFONAMIDE, NICKEL (II) ION, ... | | Authors: | Filippakopoulos, P, Eswaran, J, Keates, T, Burgess-Brown, N, Fedorov, O, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-21 | | Release date: | 2008-12-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Haspin with an Isoquinoline ligand
To be Published
|
|
3I2E
 
 | | Crystal structure of human dimethylarginine dymethylaminohydrolase-1 (DDAH-1) | | Descriptor: | N(G),N(G)-dimethylarginine dimethylaminohydrolase 1 | | Authors: | Monzingo, A.F, Wang, Y, Hu, S, Schaller, T.H, Robertus, J.D, Fast, W. | | Deposit date: | 2009-06-29 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Developing dual and specific inhibitors of dimethylarginine dimethylaminohydrolase-1 and nitric oxide synthase: toward a targeted polypharmacology to control nitric oxide.
Biochemistry, 48, 2009
|
|
3I2O
 
 | |
3I2Y
 
 | | Proteinase K by Classical hanging drop Method before high X-Ray dose on ID14-2 Beamline at ESRF | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Pechkova, E, Tripathi, S.K, Ravelli, R, McSweeney, S, Nicolini, C. | | Deposit date: | 2009-06-30 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.995 Å) | | Cite: | Atomic structure and radiation resistance of Langmuir-Blodgett protein crystals
To be Published
|
|
3HRD
 
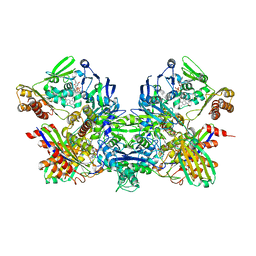 | | Crystal structure of nicotinate dehydrogenase | | Descriptor: | CALCIUM ION, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wagener, N, Pierik, A.J, Hille, R, Dobbek, H. | | Deposit date: | 2009-06-09 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Mo-Se active site of nicotinate dehydrogenase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HSS
 
 | | A higher resolution structure of Rv0554 from Mycobacterium tuberculosis complexed with malonic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Johnston, J.M, Baker, E.N. | | Deposit date: | 2009-06-10 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analysis of Rv0554 from Mycobacterium tuberculosis: testing a putative role in menaquinone biosynthesis.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3HT0
 
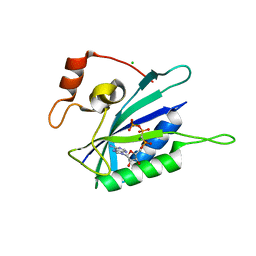 | | Crystal structure of E. coli HPPK(F123A) in complex with MgAMPCPP | | Descriptor: | CHLORIDE ION, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, HPPK, ... | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-06-11 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Pterin-binding site mutation Y53A, N55A or F123A and activity of E. coli HPPK
To be Published
|
|
3HT7
 
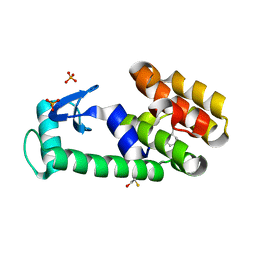 | | 2-ethylphenol in complex with T4 lysozyme L99A/M102Q | | Descriptor: | 2-ethylphenol, BETA-MERCAPTOETHANOL, Lysozyme, ... | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HTF
 
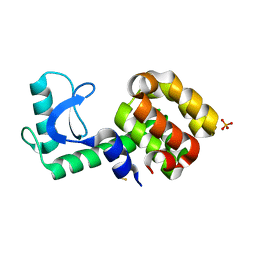 | | 4-chloro-1h-pyrazole in complex with T4 lysozyme L99A/M102Q | | Descriptor: | 4-chloro-1H-pyrazole, BETA-MERCAPTOETHANOL, Lysozyme, ... | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HTN
 
 | |
3HWS
 
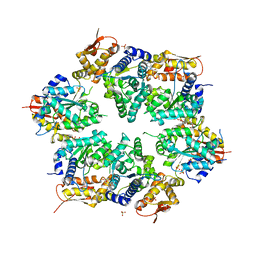 | | Crystal structure of nucleotide-bound hexameric ClpX | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit clpX, MAGNESIUM ION, ... | | Authors: | Glynn, S.E, Martin, A, Baker, T.A, Sauer, R.T. | | Deposit date: | 2009-06-18 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine.
Cell(Cambridge,Mass.), 139, 2009
|
|
3I56
 
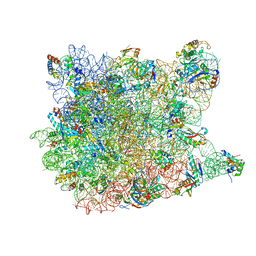 | | Co-crystal structure of Triacetyloleandomcyin Bound to the Large Ribosomal Subunit | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10E, 50S ribosomal protein L10e, ... | | Authors: | Gurel, G, Blaha, G, Steitz, T.A, Moore, P.B. | | Deposit date: | 2009-07-03 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of triacetyloleandomycin and mycalamide A bind to the large ribosomal subunit of Haloarcula marismortui.
Antimicrob.Agents Chemother., 53, 2009
|
|
3HMW
 
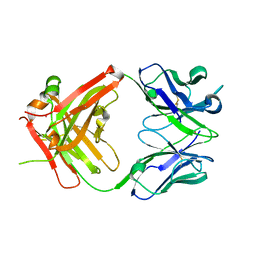 | | Crystal structure of ustekinumab FAB | | Descriptor: | CADMIUM ION, USTEKINUMAB FAB HEAVY CHAIN, USTEKINUMAB FAB LIGHT CHAIN | | Authors: | Luo, J. | | Deposit date: | 2009-05-29 | | Release date: | 2010-06-09 | | Last modified: | 2015-07-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the dual recognition of IL-12 and IL-23 by ustekinumab.
J.Mol.Biol., 402, 2010
|
|
3HYD
 
 | |
3HN7
 
 | |
3HNJ
 
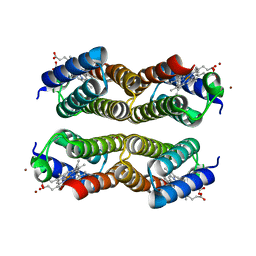 | | Crystal structure of the Zn-induced tetramer of the engineered cyt cb562 variant RIDC-2 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Soluble cytochrome b562, ZINC ION | | Authors: | Salgado, E.N, Lewis, R.A, Brodin, J, Tezcan, F.A. | | Deposit date: | 2009-05-31 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal templated design of protein interfaces.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3HZ2
 
 | |
3HNY
 
 | |
3HZB
 
 | |
3HZY
 
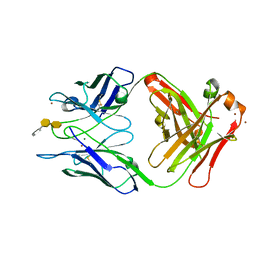 | | Crystal structure of S73-2 antibody in complex with antigen Kdo(2.4)Kdo(2.4)Kdo | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, MAGNESIUM ION, S73-2 Fab (IgG1k) heavy chain, ... | | Authors: | Brooks, C.L, Muller-Loennies, S, Borisova, S.N, Brade, L, Kosma, P, Hirama, T, MacKenzie, C.R, Brade, H, Evans, S.V. | | Deposit date: | 2009-06-24 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antibodies raised against chlamydial lipopolysaccharide antigens reveal convergence in germline gene usage and differential epitope recognition
Biochemistry, 49, 2010
|
|
3HOJ
 
 | |
3I0E
 
 | | Crystal structure of GTB C80S/C196S + H-antigen | | Descriptor: | ABO glycosyltransferase, alpha-L-fucopyranose-(1-2)-hexyl beta-D-galactopyranoside | | Authors: | Schuman, B, Persson, M, Landry, R.C, Polakowski, R, Weadge, J.T, Seto, N.O.L, Borisova, S, Palcic, M.M, Evans, S.V. | | Deposit date: | 2009-06-25 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Cysteine-to-serine mutants dramatically reorder the active site of human ABO(H) blood group B glycosyltransferase without affecting activity: structural insights into cooperative substrate binding
J.Mol.Biol., 402, 2010
|
|
3I0L
 
 | | Crystal structure of GTB C80S/C196S/C209S + DA + UDP-Gal | | Descriptor: | ABO glycosyltransferase, URIDINE-5'-DIPHOSPHATE, alpha-L-fucopyranose-(1-2)-hexyl beta-D-galactopyranoside, ... | | Authors: | Schuman, B, Persson, M, Landry, R.C, Polakowski, R, Weadge, J.T, Seto, N.O.L, Borisova, S, Palcic, M.M, Evans, S.V. | | Deposit date: | 2009-06-25 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cysteine-to-serine mutants dramatically reorder the active site of human ABO(H) blood group B glycosyltransferase without affecting activity: structural insights into cooperative substrate binding
J.Mol.Biol., 402, 2010
|
|
3I0X
 
 | |
3HQW
 
 | | Discovery of novel inhibitors of PDE10A | | Descriptor: | 4,5-bis(4-methoxyphenyl)-2-thiophen-2-yl-1H-imidazole, MAGNESIUM ION, ZINC ION, ... | | Authors: | Pandit, J, Marr, E.S. | | Deposit date: | 2009-06-08 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of a Novel Class of Phosphodiesterase 10A Inhibitors and Identification of Clinical Candidate 2-[4-(1-Methyl-4-pyridin-4-yl-1H-pyrazol-3-yl)-phenoxymethyl]-quinoline (PF-2545920) for the Treatment of Schizophrenia
J.Med.Chem., 52, 2009
|
|
