7QUE
 
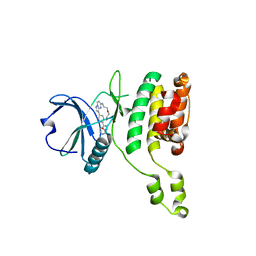 | | The STK17A (DRAK1) Kinase Domain Bound to CKJB68 | | Descriptor: | Serine/threonine-protein kinase 17A, ~{N}-(phenylmethyl)-7,10-dioxa-13,17,18,21-tetrazatetracyclo[12.5.2.1^{2,6}.0^{17,20}]docosa-1(20),2(22),3,5,14(21),15,18-heptaene-5-carboxamide | | Authors: | Mathea, S, Preuss, F, Chatterjee, D, Dederer, V, Kurz, C.G, Amrhein, J.A, Hanke, T, Knapp, S. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Illuminating the Dark: Highly Selective Inhibition of Serine/Threonine Kinase 17A with Pyrazolo[1,5- a ]pyrimidine-Based Macrocycles.
J.Med.Chem., 65, 2022
|
|
7QUF
 
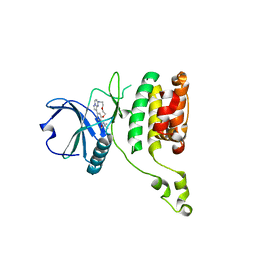 | | The STK17A (DRAK1) Kinase Domain Bound to CK156 | | Descriptor: | Serine/threonine-protein kinase 17A, ~{N}-~{tert}-butyl-7,10-dioxa-13,17,18,21-tetrazatetracyclo[12.5.2.1^{2,6}.0^{17,20}]docosa-1(20),2(22),3,5,14(21),15,18-heptaene-5-carboxamide | | Authors: | Mathea, S, Preuss, F, Chatterjee, D, Dederer, V, Kurz, C, Hanke, T, Knapp, S. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Illuminating the Dark: Highly Selective Inhibition of Serine/Threonine Kinase 17A with Pyrazolo[1,5- a ]pyrimidine-Based Macrocycles.
J.Med.Chem., 65, 2022
|
|
7QJI
 
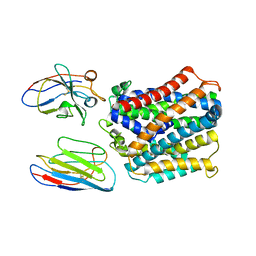 | | X-Ray Structure of apo-EleNRMT in complex with two Nanobodies at 4.1A | | Descriptor: | Divalent metal cation transporter, Elen-Nanobody-complex | | Authors: | Ramanadane, K, Straub, M.S, Dutzler, R, Manatschal, C. | | Deposit date: | 2021-12-16 | | Release date: | 2022-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural and functional properties of a magnesium transporter of the SLC11/NRAMP family.
Elife, 11, 2022
|
|
7QJJ
 
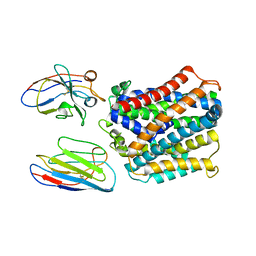 | | X-Ray Structure of a Mn2+ soak of EleNRMT in complex with two Nanobodies at 4.6A | | Descriptor: | Divalent metal cation transporter, Elen-Nb1-Nb2, MANGANESE (II) ION | | Authors: | Ramanadane, K, Straub, M.S, Dutzler, R, Manatschal, C. | | Deposit date: | 2021-12-16 | | Release date: | 2022-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Structural and functional properties of a magnesium transporter of the SLC11/NRAMP family.
Elife, 11, 2022
|
|
7L7G
 
 | | Electron cryo-microscopy of the eukaryotic translation initiation factor 2B from Homo sapiens (updated model of PDB ID: 6CAJ) | | Descriptor: | 2-(4-chloranylphenoxy)-~{N}-[4-[2-(4-chloranylphenoxy)ethanoylamino]cyclohexyl]ethanamide, Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, ... | | Authors: | Tsai, J.C, Miller-Vedam, L.E, Anand, A, Jaishankar, P, Nguyen, H.C, Wang, L, Renslo, A.R, Frost, A, Walter, P. | | Deposit date: | 2020-12-28 | | Release date: | 2021-03-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | eIF2B conformation and assembly state regulates the integrated stress response.
Elife, 10, 2021
|
|
4PPS
 
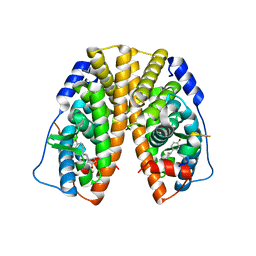 | | Crystal Structure of the Estrogen Receptor alpha Ligand-binding Domain in Complex with an A-CD ring estrogen derivative | | Descriptor: | (1S,3aR,5R,7aS)-5-(4-hydroxyphenyl)-7a-methyloctahydro-1H-inden-1-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Parent, A.A, Hughes, T.S, Pollock, J.A, Gjyshi, O, Cavett, V, Nowak, J, Garcia-Ordonez, R.D, Houtman, R, Griffin, P.R, Kojetin, D.J, Katzenellenbogen, J.A, Conkright, M.D, Nettles, K.W. | | Deposit date: | 2014-02-27 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.929 Å) | | Cite: | Resveratrol modulates the inflammatory response via an estrogen receptor-signal integration network.
Elife, 3, 2014
|
|
7Q6Y
 
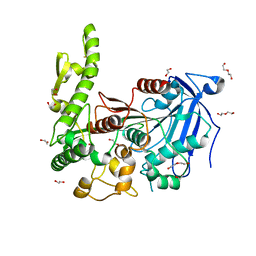 | | The X-ray crystal structure of CbTan2, a tannase enzyme from Clostridium butyricum | | Descriptor: | 1,2-ETHANEDIOL, Alpha/beta hydrolase, DI(HYDROXYETHYL)ETHER | | Authors: | Coleman, T, Mazurkewich, S, Larsbrink, J. | | Deposit date: | 2021-11-09 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural diversity and substrate preferences of three tannase enzymes encoded by the anaerobic bacterium Clostridium butyricum.
J.Biol.Chem., 298, 2022
|
|
7L48
 
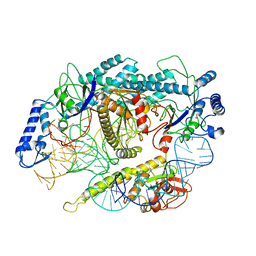 | | Cryo-EM structure of a CRISPR-Cas12f Binary Complex | | Descriptor: | Cas12f, ZINC ION, sgRNA | | Authors: | Chang, L, Li, Z. | | Deposit date: | 2020-12-18 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for substrate recognition and cleavage by the dimerization-dependent CRISPR-Cas12f nuclease.
Nucleic Acids Res., 49, 2021
|
|
4PPP
 
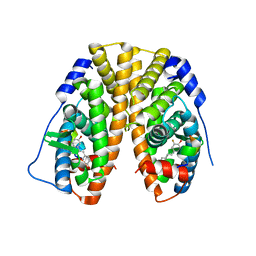 | | Crystal Structure of the Estrogen Receptor alpha Ligand-binding Domain in Complex with Fluoro-Resveratrol | | Descriptor: | 5-[(E)-2-(3-fluoro-4-hydroxyphenyl)ethenyl]benzene-1,3-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Parent, A.A, Hughes, T.S, Pollock, J.A, Gjyshi, O, Cavett, V, Nowak, J, Garcia-Ordonez, R.D, Houtman, R, Griffin, P.R, Kojetin, D.J, Katzenellenbogen, J.A, Conkright, M.D, Nettles, K.W. | | Deposit date: | 2014-02-27 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.686 Å) | | Cite: | Resveratrol modulates the inflammatory response via an estrogen receptor-signal integration network.
Elife, 3, 2014
|
|
4PR8
 
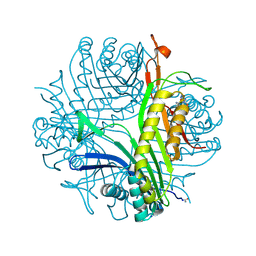 | |
7QBZ
 
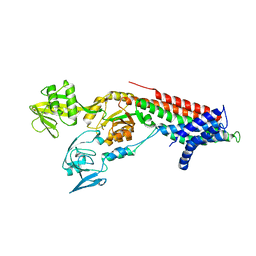 | | Crystal structure Cadmium translocating P-type ATPase | | Descriptor: | Cadmium translocating P-type ATPase, MAGNESIUM ION, TETRAFLUOROALUMINATE ION | | Authors: | Groenberg, C, Hu, Q, Wang, K, Gourdon, P. | | Deposit date: | 2021-11-21 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure and ion-release mechanism of P IB-4 -type ATPases.
Elife, 10, 2021
|
|
7QC0
 
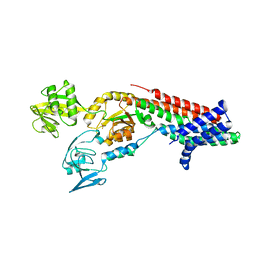 | | Crystal structure of Cadmium translocating P-type ATPase | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Cadmium translocating P-type ATPase, MAGNESIUM ION | | Authors: | Groenberg, C, Hu, Q, Wang, K, Gourdon, P. | | Deposit date: | 2021-11-21 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structure and ion-release mechanism of P IB-4 -type ATPases.
Elife, 10, 2021
|
|
4Q3M
 
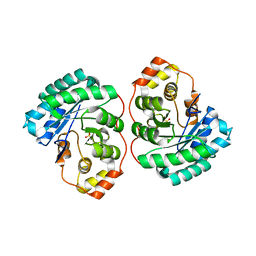 | | Crystal structure of MGS-M4, an aldo-keto reductase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | MGS-M4, SODIUM ION, SULFATE ION | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.552 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
6TU3
 
 | | Rat 20S proteasome | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Deshmukh, F.K, Polkinghorn, C.R, Elad, N, Sharon, M. | | Deposit date: | 2020-01-02 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Comparative Structural Analysis of 20S Proteasome Ortholog Protein Complexes by Native Mass Spectrometry.
Acs Cent.Sci., 6, 2020
|
|
7QUR
 
 | | SARS-CoV-2 Spike with ethylbenzamide-tri-iodo Siallyllactose, C3 symmetry | | Descriptor: | 2,3,5-tris(iodanyl)benzamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Naismith, J.H, Yang, Y, Liu, J.W. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-01 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Pathogen-sugar interactions revealed by universal saturation transfer analysis.
Science, 377, 2022
|
|
7QUS
 
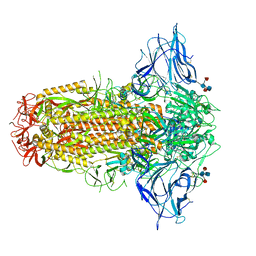 | | SARS-CoV-2 Spike, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Naismith, J.H, Yang, Y, Liu, J.W. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-08 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Pathogen-sugar interactions revealed by universal saturation transfer analysis.
Science, 377, 2022
|
|
6NJM
 
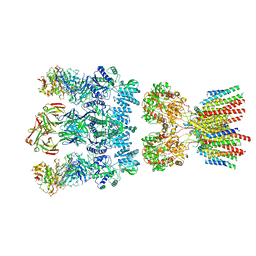 | | Architecture and subunit arrangement of native AMPA receptors | | Descriptor: | 15F1 Fab heavy chain, 15F1 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gouaux, E, Zhao, Y. | | Deposit date: | 2019-01-03 | | Release date: | 2019-04-24 | | Last modified: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Architecture and subunit arrangement of native AMPA receptors elucidated by cryo-EM.
Science, 364, 2019
|
|
4Q3K
 
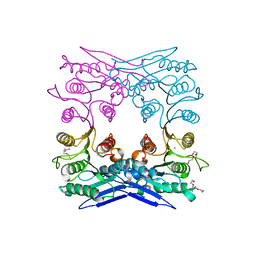 | | Crystal structure of MGS-M1, an alpha/beta hydrolase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | CHLORIDE ION, FLUORIDE ION, MGS-M1, ... | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
1C2X
 
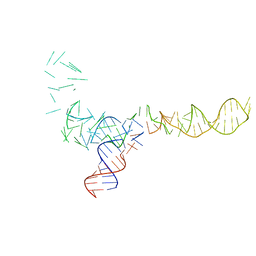 | |
4Q3L
 
 | | Crystal structure of MGS-M2, an alpha/beta hydrolase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | GLYCEROL, MGS-M2 | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
4Q1U
 
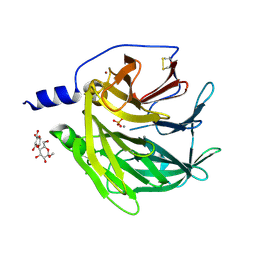 | | Serum paraoxonase-1 by directed evolution with the K192Q mutation | | Descriptor: | BROMIDE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ben-David, M, Sussman, J.L, Tawfik, D.S. | | Deposit date: | 2014-04-04 | | Release date: | 2015-02-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Catalytic stimulation by restrained active-site floppiness-the case of high density lipoprotein-bound serum paraoxonase-1.
J.Mol.Biol., 427, 2015
|
|
6NJL
 
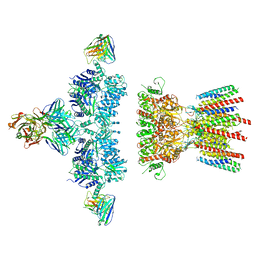 | |
7Q94
 
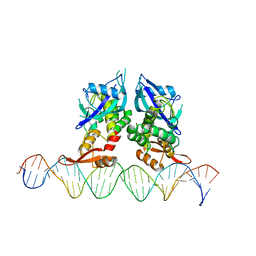 | | Crystal Structure of Agrobacterium tumefaciens NADQ, DNA complex. | | Descriptor: | DNA binding region (31-MER), NADQ transcription factor | | Authors: | Cianci, M, Minazzato, G, Heroux, A, Raffaelli, N, Sorci, L, Gasparrini, M. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Bacterial NadQ (COG4111) is a Nudix-like, ATP-responsive regulator of NAD biosynthesis.
J.Struct.Biol., 214, 2022
|
|
7Q91
 
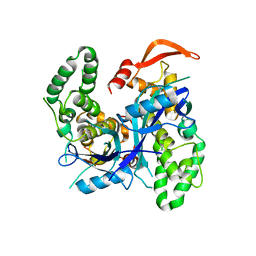 | | Crystal Structure of Agrobacterium tumefaciens NADQ, native form. | | Descriptor: | NADQ transcription factor, SODIUM ION | | Authors: | Cianci, M, Minazzato, G, Heroux, A, Raffaelli, N, Sorci, L, Gasparrini, M. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Bacterial NadQ (COG4111) is a Nudix-like, ATP-responsive regulator of NAD biosynthesis.
J.Struct.Biol., 214, 2022
|
|
7Q93
 
 | | Crystal Structure of Agrobacterium tumefaciens NADQ, NAD complex. | | Descriptor: | GLYCEROL, NADQ transcription factor, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Cianci, M, Minazzato, G, Heroux, A, Raffaelli, N, Sorci, L, Gasparrini, M. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Bacterial NadQ (COG4111) is a Nudix-like, ATP-responsive regulator of NAD biosynthesis.
J.Struct.Biol., 214, 2022
|
|
