3DJT
 
 | |
3DK0
 
 | |
3DYD
 
 | | Human Tyrosine Aminotransferase | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Tyrosine aminotransferase | | Authors: | Karlberg, T, Moche, M, Andersson, J, Arrowsmith, C.H, Berglund, H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kotenyova, T, Lehtio, L, Nilsson, M.E, Nordlund, P, Nyman, T, Olesen, K, Persson, C, Sagemark, J, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wikstrom, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-07-27 | | Release date: | 2008-08-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human Tyrosine Aminotransferase
To be Published
|
|
2F8J
 
 | |
3DK2
 
 | |
2JOX
 
 | | Embryonic Neural Inducing Factor Churchill is not a DNA-Binding Zinc Finger Protein: Solution Structure Reveals a Solvent-Exposed beta-Sheet and Zinc Binuclear Cluster | | Descriptor: | Churchill protein, ZINC ION | | Authors: | Lee, B.M, Buck-Koehntop, B.A, Martinez-Yamout, M.A, Gottesfeld, J.M, Dyson, H, Wright, P.E. | | Deposit date: | 2007-04-07 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Embryonic Neural Inducing Factor Churchill Is not a DNA-binding Zinc Finger Protein: Solution Structure Reveals a Solvent-exposed beta-Sheet and Zinc Binuclear Cluster
J.Mol.Biol., 371, 2007
|
|
2GB3
 
 | |
3CL7
 
 | | Crystal structure of Puue Allantoinase in complex with Hydantoin | | Descriptor: | Puue Allantoinase, imidazolidine-2,4-dione | | Authors: | Ramazzina, I, Cendron, L, Folli, C, Berni, R, Monteverdi, D, Zanotti, G, Percudani, R. | | Deposit date: | 2008-03-18 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Logical identification of an allantoinase analog (puuE) recruited from polysaccharide deacetylases
J.Biol.Chem., 283, 2008
|
|
3CL8
 
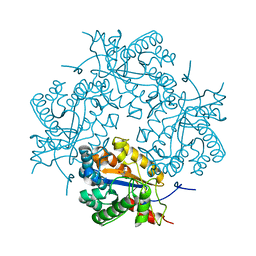 | | Crystal structure of Puue Allantoinase complexed with ACA | | Descriptor: | 5-amino-1H-imidazole-4-carboxamide, Puue Allantoinase | | Authors: | Ramazzina, I, Cendron, L, Folli, C, Berni, R, Monteverdi, D, Zanotti, G, Percudani, R. | | Deposit date: | 2008-03-18 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Logical identification of an allantoinase analog (puuE) recruited from polysaccharide deacetylases
J.Biol.Chem., 283, 2008
|
|
2B4N
 
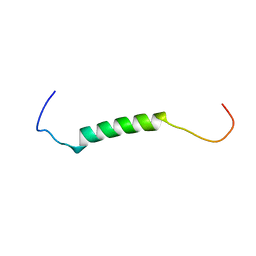 | |
3CL6
 
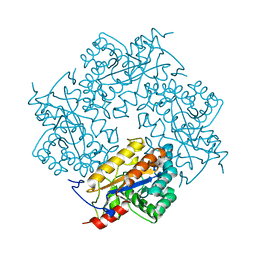 | | Crystal structure of Puue Allantoinase | | Descriptor: | Puue allantoinase | | Authors: | Ramazzina, I, Cendron, L, Folli, C, Berni, R, Monteverdi, D, Zanotti, G, Percudani, R. | | Deposit date: | 2008-03-18 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Logical identification of an allantoinase analog (puuE) recruited from polysaccharide deacetylases
J.Biol.Chem., 283, 2008
|
|
2K4D
 
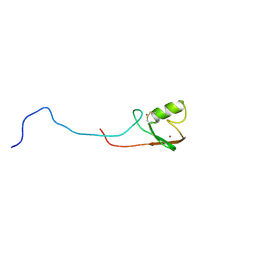 | | E2-c-Cbl recognition is necessary but not sufficient for ubiquitination activity | | Descriptor: | E3 ubiquitin-protein ligase CBL, ZINC ION | | Authors: | Huang, A, De Jong, R.N, Wienk, H, Winkler, S.G, Timmers, H.T.M, Boelens, R. | | Deposit date: | 2008-06-06 | | Release date: | 2009-01-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | E2-c-Cbl recognition is necessary but not sufficient for ubiquitination activity
J.Mol.Biol., 385, 2009
|
|
2Z61
 
 | |
4WMA
 
 | |
2DOU
 
 | | probable N-succinyldiaminopimelate aminotransferase (TTHA0342) from Thermus thermophilus HB8 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION, probable N-succinyldiaminopimelate aminotransferase | | Authors: | Omi, R, Goto, M, Miyahara, I, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-05-03 | | Release date: | 2006-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | probable N-succinyldiaminopimelate aminotransferase (TTHA0342) from Thermus thermophilus HB8
To be published
|
|
4WM0
 
 | |
4WN2
 
 | |
2GSA
 
 | |
1M6F
 
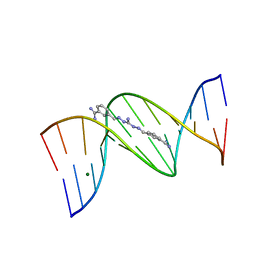 | | Strong Binding in the DNA Minor Groove by an Aromatic Diamidine With a Shape That Does Not Match the Curvature of the Groove | | Descriptor: | 3-[C-[N'-(3-CARBAMIMIDOYL-BENZYLIDENIUM)-HYDRAZINO]-[[AMINOMETHYLIDENE]AMINIUM]-IMINOMETHYL]-BENZAMIDINIUM, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Nguyen, B, Lee, M.P.H, Hamelberg, D, Joubert, A, Bailly, C, Brun, R, Neidle, S, Wilson, W.D. | | Deposit date: | 2002-07-16 | | Release date: | 2002-11-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Strong Binding in the DNA Minor Groove by an Aromatic Diamidine With a Shape That Does Not Match the Curvature of the Groove
J.Am.Chem.Soc., 124, 2002
|
|
1B44
 
 | | CRYSTAL STRUCTURE OF THE B SUBUNIT OF HEAT-LABILE ENTEROTOXIN FROM E. COLI CARRYING A PEPTIDE WITH ANTI-HSV ACTIVITY | | Descriptor: | PROTEIN (B-POL SUBUNIT OF HEAT-LABILE ENTEROTOXIN) | | Authors: | Matkovic-Calogovic, D, Loregian, A, D'Acunto, M.R, Battistutta, R, Tossi, A, Palu, G, Zanotti, G. | | Deposit date: | 1999-01-04 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the B subunit of Escherichia coli heat-labile enterotoxin carrying peptides with anti-herpes simplex virus type 1 activity.
J.Biol.Chem., 274, 1999
|
|
4WBT
 
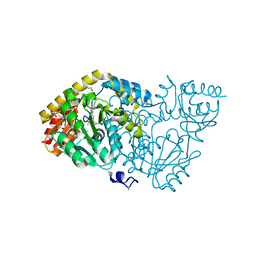 | | Crystal structure of histidinol-phosphate aminotransferase from Sinorhizobium meliloti in complex with pyridoxal-5'-phosphate | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Shabalin, I.G, Bacal, P, Kowalska, A.K, Cooper, D.R, Stead, M, Hammonds, J, Ahmed, M, Hillerich, B.S, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-03 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of histidinol-phosphate aminotransferase from Sinorhizobium meliloti in complex with pyridoxal-5'-phosphate
to be published
|
|
3DMW
 
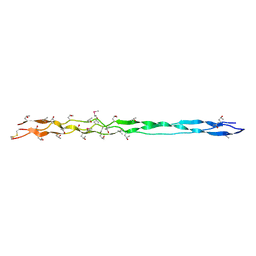 | | Crystal structure of human type III collagen G982-G1023 containing C-terminal cystine knot | | Descriptor: | Collagen alpha-1(III) chain | | Authors: | Boudko, S.P, Engel, J, Okuyama, K, Mizuno, K, Bachinger, H.P, Schumacher, M.A. | | Deposit date: | 2008-07-01 | | Release date: | 2008-09-30 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human type III collagen Gly991-Gly1032 cystine knot-containing peptide shows both 7/2 and 10/3 triple helical symmetries.
J.Biol.Chem., 283, 2008
|
|
3DO4
 
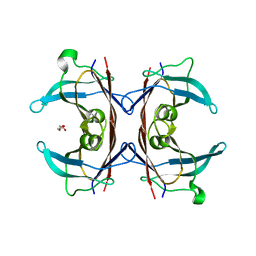 | |
2PBW
 
 | | Crystal Structure of the Ligand-Binding Core of iGluR5 in Complex with the Partial agonist Domoic Acid at 2.5 A Resolution | | Descriptor: | (2S,3S,4S)-2-CARBOXY-4-[(1Z,3E,5R)-5-CARBOXY-1-METHYL-1,3-HEXADIENYL]-3-PYRROLIDINEACETIC ACID, Glutamate receptor, ionotropic kainate 1 | | Authors: | Hald, H, Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-03-29 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Partial agonism and antagonism of the ionotropic glutamate receptor iGLuR5: structures of the ligand-binding core in complex with domoic acid and 2-amino-3-[5-tert-butyl-3-(phosphonomethoxy)-4-isoxazolyl]propionic acid.
J.Biol.Chem., 282, 2007
|
|
2G3V
 
 | | Crystal structure of CagS (HP0534, Cag13) from Helicobacter pylori | | Descriptor: | (UNK)(UNK)(UNK)(UNK)(UNK)(MSE)(UNK), CAG pathogenicity island protein 13 | | Authors: | Cendron, L, Tasca, E, Angelini, A, Seydel, A, Battistutta, R, Montecucco, C, Zanotti, G. | | Deposit date: | 2006-02-21 | | Release date: | 2007-03-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of CagS from helicobacter pylori
To be Published
|
|
