7TF5
 
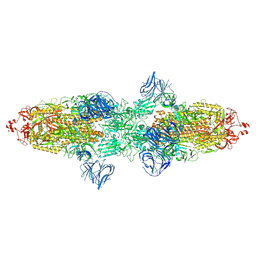 | | Cryo-EM structure of SARS-CoV-2 Kappa (B.1.617.1) spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural and biochemical rationale for enhanced spike protein fitness in delta and kappa SARS-CoV-2 variants.
Nat Commun, 13, 2022
|
|
4M9Y
 
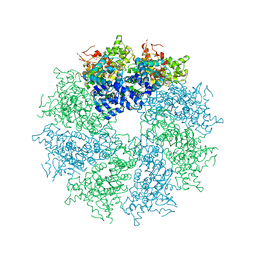 | | Crystal structure of CED-4 bound CED-3 fragment | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CED-3 fragment, Cell death protein 4, ... | | Authors: | Huang, W.J, Jinag, T.Y, Choi, W.Y, Wang, J.W, Shi, Y.G. | | Deposit date: | 2013-08-15 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Mechanistic insights into CED-4-mediated activation of CED-3.
Genes Dev., 27, 2013
|
|
3CSJ
 
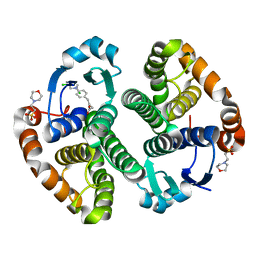 | | Human glutathione s-transferase p1-1 in complex with chlorambucil | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORAMBUCIL, CHLORIDE ION, ... | | Authors: | Parker, L.J. | | Deposit date: | 2008-04-09 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The anti-cancer drug chlorambucil as a substrate for the human polymorphic enzyme glutathione transferase P1-1: kinetic properties and crystallographic characterisation of allelic variants.
J.Mol.Biol., 380, 2008
|
|
4FXM
 
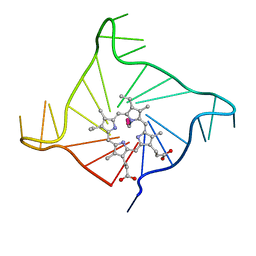 | | Crystal structure of the complex of a human telomeric repeat G-quadruplex and N-methyl mesoporphyrin IX (P21212) | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3'), N-METHYLMESOPORPHYRIN, POTASSIUM ION | | Authors: | Nicoludis, J.M, Miller, S.T, Jeffrey, P, Lawton, T.J, Rosenzweig, A.C, Yatsunyk, L.A. | | Deposit date: | 2012-07-03 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Optimized End-Stacking Provides Specificity of N-Methyl Mesoporphyrin IX for Human Telomeric G-Quadruplex DNA.
J.Am.Chem.Soc., 134, 2012
|
|
1TZI
 
 | |
5W88
 
 | |
5WCL
 
 | | NMR structure of the N-domain of troponin C bound to switch region of troponin I and 3-methyldiphenylamine (solvent exposed mode) | | Descriptor: | 3-methyl-N-phenylaniline, Troponin C, slow skeletal and cardiac muscles,Troponin I, ... | | Authors: | Cai, F, Hwang, P.M, Sykes, B.D. | | Deposit date: | 2017-06-30 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures reveal details of small molecule binding to cardiac troponin.
J. Mol. Cell. Cardiol., 101, 2016
|
|
4FYC
 
 | | Structural and functional characterizations of a thioredoxin-fold protein from Helicobacter pylori | | Descriptor: | TETRAETHYLENE GLYCOL, Thiol:disulfide interchange protein (DsbC) | | Authors: | Yoon, J.Y, Kim, J, Lee, S.J, Im, H.N, Kim, H.S, Yoon, H, An, D.R, Kim, J.Y, Kim, S, Han, B.W, Suh, S.W. | | Deposit date: | 2012-07-04 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural and functional characterization of HP0377, a thioredoxin-fold protein from Helicobacter pylori
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7SV8
 
 | |
7TF4
 
 | | Cryo-EM structure of SARS-CoV-2 Kappa (B.1.617.1) spike protein (focused refinement of RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural and biochemical rationale for enhanced spike protein fitness in delta and kappa SARS-CoV-2 variants.
Nat Commun, 13, 2022
|
|
7T9H
 
 | | HIV Integrase in complex with Compound-15 | | Descriptor: | (2S)-tert-butoxy[2-methyl-4-(4-methylphenyl)quinolin-3-yl]acetic acid, Integrase, MAGNESIUM ION | | Authors: | Khan, J.A, Lewis, H, Kish, K. | | Deposit date: | 2021-12-19 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Design, Synthesis, and Preclinical Profiling of GSK3739936 (BMS-986180), an Allosteric Inhibitor of HIV-1 Integrase with Broad-Spectrum Activity toward 124/125 Polymorphs.
J.Med.Chem., 65, 2022
|
|
3CUE
 
 | | Crystal structure of a TRAPP subassembly activating the Rab Ypt1p | | Descriptor: | GTP-binding protein YPT1, PALMITIC ACID, Transport protein particle 18 kDa subunit, ... | | Authors: | Cai, Y, Reinisch, K.M. | | Deposit date: | 2008-04-16 | | Release date: | 2008-07-08 | | Last modified: | 2012-03-21 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | The structural basis for activation of the Rab Ypt1p by the TRAPP membrane-tethering complexes.
Cell(Cambridge,Mass.), 133, 2008
|
|
4FZJ
 
 | | Pantothenate synthetase in complex with 1,3-DIMETHYL-1H-THIENO[2,3-C]PYRAZOLE-5-CARBOXYLIC ACID | | Descriptor: | 1,2-ETHANEDIOL, 1,3-dimethyl-1H-thieno[2,3-c]pyrazole-5-carboxylic acid, ETHANOL, ... | | Authors: | Silvestre, H.L, Blundell, T.L, Abell, C, Ciulli, A. | | Deposit date: | 2012-07-06 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Integrated biophysical approach to fragment screening and validation for fragment-based lead discovery.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4FZZ
 
 | | Exonuclease X in complex with 5' overhanging duplex DNA | | Descriptor: | DNA (5'-D(*GP*TP*CP*AP*TP*TP*GP*TP*GP*GP*AP*TP*CP*CP*GP*AP*G)-3'), Exodeoxyribonuclease 10, SODIUM ION | | Authors: | Wang, T, Sun, H, Cheng, F, Bi, L, Jiang, T. | | Deposit date: | 2012-07-08 | | Release date: | 2013-07-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recognition and processing of double-stranded DNA by ExoX, a distributive 3'-5' exonuclease
Nucleic Acids Res., 41, 2013
|
|
3CZT
 
 | | Crystal Structure of S100B in the Calcium and Zinc Loaded State at pH 9 | | Descriptor: | CALCIUM ION, Protein S100-B, ZINC ION | | Authors: | Ostendorp, T, Diez, J, Heizmann, C.W, Fritz, G. | | Deposit date: | 2008-04-30 | | Release date: | 2009-04-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The crystal structures of human S100B in the zinc- and calcium-loaded state at three pH values reveal zinc ligand swapping.
Biochim.Biophys.Acta, 1813, 2011
|
|
2RV3
 
 | | Solution structures of the DNA-binding domain (ZF15) of immune-related zinc-finger protein ZFAT | | Descriptor: | ZINC ION, Zinc finger protein ZFAT | | Authors: | Tochio, N, Umehara, T, Kigawa, T, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT
J.Struct.Funct.Genom., 16, 2015
|
|
7SUW
 
 | |
4FVK
 
 | | Structural and functional characterization of neuraminidase-like molecule N10 derived from bat influenza A virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Li, Q, Sun, X.M, Li, Z.X, Liu, Y, Vavricka, C.J, Qi, J.X, Gao, G.F. | | Deposit date: | 2012-06-29 | | Release date: | 2012-09-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structural and functional characterization of neuraminidase-like molecule N10 derived from bat influenza A virus
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3D0Y
 
 | | Crystal Structure of S100B in the Calcium and Zinc Loaded State at pH 6.5 | | Descriptor: | CALCIUM ION, Protein S100-B, TETRAETHYLENE GLYCOL, ... | | Authors: | Ostendorp, T, Diez, J, Heizmann, C.W, Fritz, G. | | Deposit date: | 2008-05-02 | | Release date: | 2009-04-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structures of human S100B in the zinc- and calcium-loaded state at three pH values reveal zinc ligand swapping.
Biochim.Biophys.Acta, 1813, 2011
|
|
2SPG
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T15S | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-21 | | Release date: | 1999-04-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
3D1H
 
 | |
7SV1
 
 | |
4G0F
 
 | | Crystal structure of the complex of a human telomeric repeat G-quadruplex and N-methyl mesoporphyrin IX (P6) | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3'), N-METHYLMESOPORPHYRIN, POTASSIUM ION | | Authors: | Nicoludis, J.M, Miller, S.T, Jeffrey, P, Lawton, T.J, Rosenzweig, A.C, Yatsunyk, L.A. | | Deposit date: | 2012-07-09 | | Release date: | 2012-12-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Optimized End-Stacking Provides Specificity of N-Methyl Mesoporphyrin IX for Human Telomeric G-Quadruplex DNA.
J.Am.Chem.Soc., 134, 2012
|
|
2RK2
 
 | | DHFR R-67 complexed with NADP | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Dihydrofolate reductase type 2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Krahn, J.M, London, R.E. | | Deposit date: | 2007-10-16 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a type II dihydrofolate reductase catalytic ternary complex.
Biochemistry, 46, 2007
|
|
3D4V
 
 | | Crystal Structure of an AlkA Host/Guest Complex N7MethylGuanine:Cytosine Base Pair | | Descriptor: | 5'-D(*DGP*DAP*DCP*DAP*DTP*DGP*DAP*(FMG)P*DTP*DGP*DCP*DC)-3', 5'-D(*DGP*DGP*DCP*DAP*DCP*DTP*DCP*DAP*DTP*DGP*DTP*DC)-3', DNA-3-methyladenine glycosylase 2 | | Authors: | Lee, S, Bowman, B.R, Wang, S, Verdine, G.L. | | Deposit date: | 2008-05-15 | | Release date: | 2008-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Synthesis and structure of duplex DNA containing the genotoxic nucleobase lesion N7-methylguanine.
J.Am.Chem.Soc., 130, 2008
|
|
