7SZE
 
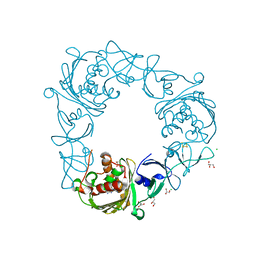 | |
3D4C
 
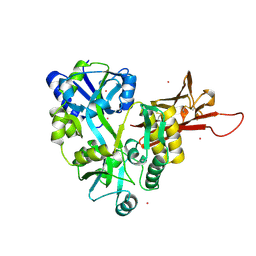 | |
4E0F
 
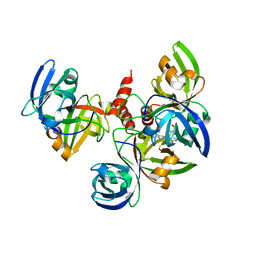 | | Crystallographic structure of trimeric Riboflavin Synthase from Brucella abortus in complex with riboflavin | | Descriptor: | RIBOFLAVIN, Riboflavin synthase subunit alpha | | Authors: | Serer, M.I, Bonomi, H.R, Guimaraes, B.G, Rossi, R.C, Goldbaum, F.A, Klinke, S. | | Deposit date: | 2012-03-03 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystallographic and kinetic study of riboflavin synthase from Brucella abortus, a chemotherapeutic target with an enhanced intrinsic flexibility.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7SZH
 
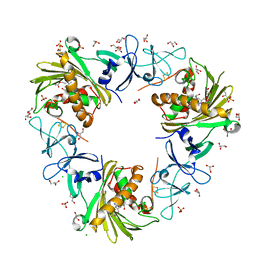 | |
1E52
 
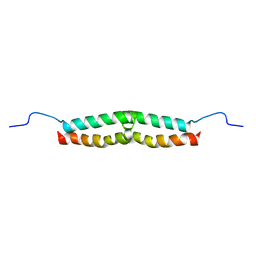 | | Solution structure of Escherichia coli UvrB C-terminal domain | | Descriptor: | EXCINUCLEASE ABC SUBUNIT | | Authors: | Alexandrovich, A.A, Kelly, G.G, Frenkiel, T.A, Moolenaar, G.F, Goosen, N.N, Sanderson, M.R, Lane, A.N. | | Deposit date: | 2000-07-14 | | Release date: | 2001-07-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution Structure, Hydrodynamics and Thermodynamics of the Uvrb C-Terminal Domain.
J.Biomol.Struct.Dyn., 19, 2001
|
|
3D78
 
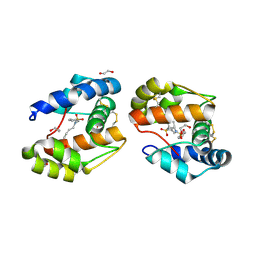 | | Dimeric crystal structure of a pheromone binding protein mutant D35N, from apis mellifera, at pH 7.0 | | Descriptor: | 1,2-ETHANEDIOL, N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-05-20 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
2RSY
 
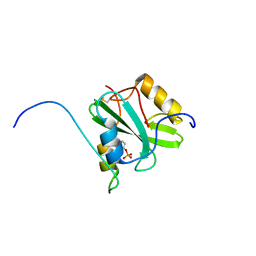 | | Solution structure of the SH2 domain of Csk in complex with a phosphopeptide from Cbp | | Descriptor: | Phosphoprotein associated with glycosphingolipid-enriched microdomains 1, Tyrosine-protein kinase CSK | | Authors: | Tanaka, H, Akagi, K, Oneyama, C, Tanaka, M, Sasaki, Y, Kanou, T, Lee, Y, Yokogawa, D, Debenecker, M, Nakagawa, A, Okada, M, Ikegami, T. | | Deposit date: | 2012-09-10 | | Release date: | 2013-04-10 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Identification of a new interaction mode between the Src homology 2 domain of C-terminal Src kinase (Csk) and Csk-binding protein/phosphoprotein associated with glycosphingolipid microdomains.
J.Biol.Chem., 288, 2013
|
|
2RUX
 
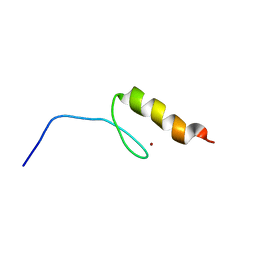 | | Solution structures of the DNA-binding domain (ZF6) of immune-related zinc-finger protein ZFAT | | Descriptor: | ZINC ION, Zinc finger protein ZFAT | | Authors: | Tochio, N, Umehara, T, Kigawa, T, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT
J.Struct.Funct.Genom., 16, 2015
|
|
2RDS
 
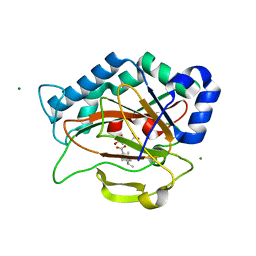 | | Crystal Structure of PtlH with Fe/oxalylglycine and ent-1-deoxypentalenic acid bound | | Descriptor: | (1S,3aS,5aR,8aS)-1,7,7-trimethyl-1,2,3,3a,5a,6,7,8-octahydrocyclopenta[c]pentalene-4-carboxylic acid, 1-deoxypentalenic acid 11-beta hydroxylase; Fe(II)/alpha-ketoglutarate dependent hydroxylase, FE (III) ION, ... | | Authors: | You, Z, Omura, S, Ikeda, H, Cane, D.E, Jogl, G. | | Deposit date: | 2007-09-24 | | Release date: | 2007-10-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the Non-heme Iron Dioxygenase PtlH in Pentalenolactone Biosynthesis.
J.Biol.Chem., 282, 2007
|
|
2RV4
 
 | | Solution structures of the DNA-binding domain (ZF5) of mouse immune-related zinc-finger protein ZFAT | | Descriptor: | ZINC ION, Zinc finger protein ZFAT | | Authors: | Tochio, N, Umehara, T, Kigawa, T, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT
J.Struct.Funct.Genom., 16, 2015
|
|
7T1V
 
 | |
7SZG
 
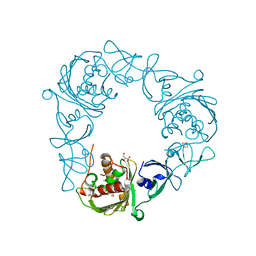 | | Structure of the Rieske Non-heme Iron Oxygenase GxtA Pressurized with Xenon | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, FE (III) ION, ... | | Authors: | Bridwell-Rabb, J, Liu, J. | | Deposit date: | 2021-11-27 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Design principles for site-selective hydroxylation by a Rieske oxygenase.
Nat Commun, 13, 2022
|
|
7SZF
 
 | |
2TSA
 
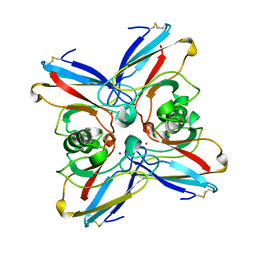 | | AZURIN MUTANT M121A | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Tsai, L.-C, Bonander, N, Harata, K, Karlsson, B.G, Vanngard, T, Langer, V, Sjolin, L. | | Deposit date: | 1996-05-10 | | Release date: | 1996-11-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutant Met121Ala of Pseudomonas aeruginosa azurin and its azide derivative: crystal structures and spectral properties.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
4FZD
 
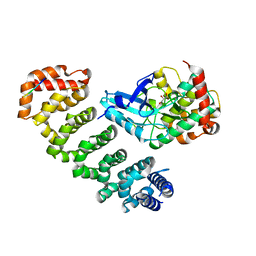 | | Crystal structure of MST4-MO25 complex with WSF motif | | Descriptor: | C-terminal peptide from Serine/threonine-protein kinase MST4, Calcium-binding protein 39, GLYCEROL, ... | | Authors: | Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2012-07-06 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of the MST4 in Complex with MO25 Provides Insights into Its Activation Mechanism
Structure, 21, 2013
|
|
4FZY
 
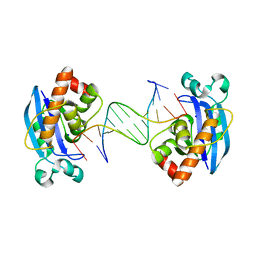 | | Exonuclease X in complex with 12bp blunt-ended dsDNA | | Descriptor: | DNA (5'-D(*TP*GP*TP*AP*GP*AP*TP*TP*CP*GP*AP*G)-3'), DNA (5'-D(P*CP*TP*CP*GP*AP*AP*TP*CP*TP*AP*CP*A)-3'), Exodeoxyribonuclease 10, ... | | Authors: | Wang, T, Sun, H, Cheng, F, Bi, L, Jiang, T. | | Deposit date: | 2012-07-08 | | Release date: | 2013-07-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Recognition and processing of double-stranded DNA by ExoX, a distributive 3'-5' exonuclease
Nucleic Acids Res., 41, 2013
|
|
1PCP
 
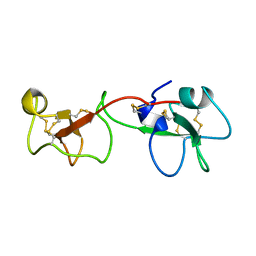 | | SOLUTION STRUCTURE OF A TREFOIL-MOTIF-CONTAINING CELL GROWTH FACTOR, PORCINE SPASMOLYTIC PROTEIN | | Descriptor: | PORCINE SPASMOLYTIC PROTEIN | | Authors: | Carr, M.D, Bauer, C.J, Gradwell, M.J, Feeney, J. | | Deposit date: | 1993-02-04 | | Release date: | 1994-05-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a trefoil-motif-containing cell growth factor, porcine spasmolytic protein.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
2RI1
 
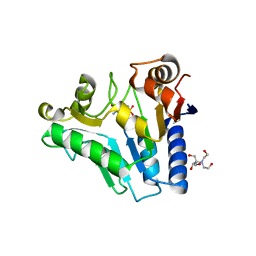 | | Crystal Structure of glucosamine 6-phosphate deaminase (NagB) with GlcN6P from S. mutans | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, Glucosamine-6-phosphate deaminase | | Authors: | Liu, C, Li, D, Su, X.D. | | Deposit date: | 2007-10-10 | | Release date: | 2008-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Ring-opening mechanism revealed by crystal structures of NagB and its ES intermediate complex
J.Mol.Biol., 379, 2008
|
|
3D5R
 
 | | Crystal Structure of Efb-C (N138A) / C3d Complex | | Descriptor: | Complement C3, Fibrinogen-binding protein | | Authors: | Geisbrecht, B.V. | | Deposit date: | 2008-05-16 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Electrostatic contributions drive the interaction between Staphylococcus aureus protein Efb-C and its complement target C3d.
Protein Sci., 17, 2008
|
|
4M9X
 
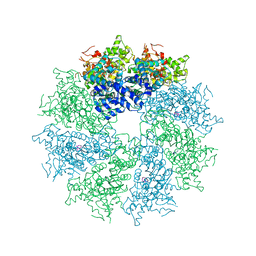 | | Crystal structure of CED-4 bound CED-3 fragment | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CED-3 fragment, Cell death protein 4, ... | | Authors: | Huang, W.J, Jinag, T.Y, Choi, W.Y, Wang, J.W, Shi, Y.G. | | Deposit date: | 2013-08-15 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.344 Å) | | Cite: | Mechanistic insights into CED-4-mediated activation of CED-3.
Genes Dev., 27, 2013
|
|
4G37
 
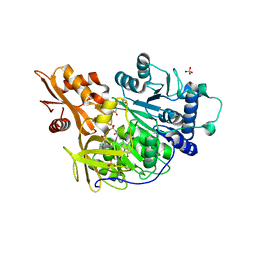 | | Structure of cross-linked firefly luciferase in second catalytic conformation | | Descriptor: | 4,4'-(ethylenediimino)bis[4-oxobutyrate], 5'-O-[N-(DEHYDROLUCIFERYL)-SULFAMOYL] ADENOSINE, CHLORIDE ION, ... | | Authors: | Sundlov, J.A, Branchini, B.R, Gulick, A.M. | | Deposit date: | 2012-07-13 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Crystal structure of firefly luciferase in a second catalytic conformation supports a domain alternation mechanism.
Biochemistry, 51, 2012
|
|
3D73
 
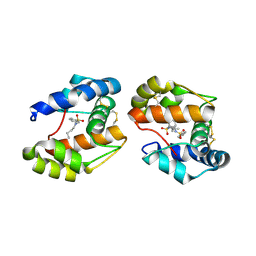 | | Crystal structure of a pheromone binding protein mutant D35A, from Apis mellifera, at pH 7.0 | | Descriptor: | N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-05-20 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
4G3V
 
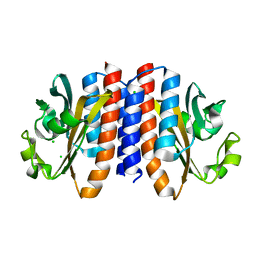 | | Crystal structure of A. Aeolicus nlh2 gaf domain in an inactive state | | Descriptor: | CHLORIDE ION, Transcriptional regulator nlh2 | | Authors: | Batchelor, J.D, Lee, P, Wang, A, Doucleff, M, Wemmer, D.E. | | Deposit date: | 2012-07-15 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural mechanism of GAF-regulated delta(54) activators from Aquifex aeolicus
J.Mol.Biol., 425, 2013
|
|
3CFB
 
 | |
4G61
 
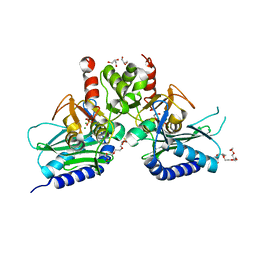 | | Crystal structure of IMPase/NADP phosphatase complexed with Mg2+ and phosphate | | Descriptor: | 1-HYDROXYSULFANYL-4-MERCAPTO-BUTANE-2,3-DIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, ... | | Authors: | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | Deposit date: | 2012-07-18 | | Release date: | 2013-07-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural elucidation of the binding site and mode of inhibition of Li(+) and Mg(2+) in inositol monophosphatase.
Febs J., 281, 2014
|
|
