2RIO
 
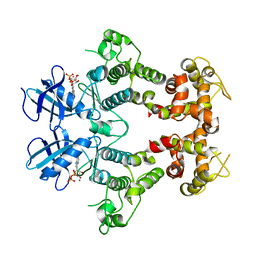 | | Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation of non-conventional splicing | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, STRONTIUM ION, ... | | Authors: | Lee, K.P, Dey, M, Neculai, D, Cao, C, Dever, T.E, Sicheri, F. | | Deposit date: | 2007-10-12 | | Release date: | 2008-01-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the dual enzyme ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing.
Cell(Cambridge,Mass.), 132, 2008
|
|
2R5J
 
 | |
4U5A
 
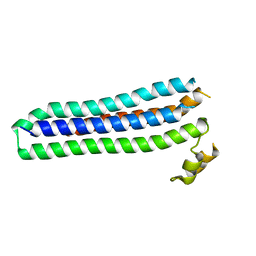 | | Sporozoite Protein for Cell Traversal | | Descriptor: | Sporozoite microneme protein essential for cell traversal | | Authors: | Hamaoka, B.Y. | | Deposit date: | 2014-07-25 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of the Essential Plasmodium Host Cell Traversal Protein SPECT1.
Plos One, 9, 2014
|
|
9EUP
 
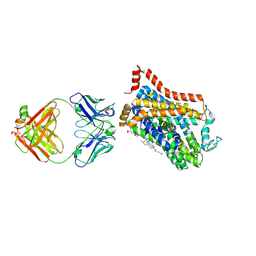 | | Inhibitor-free outward-open structure of Drosophila dopamine transporter | | Descriptor: | 9D5 ANTIBODY, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Pedersen, C.N, Yang, F, Ita, S, Xu, Y, Akunuri, R, Trampari, S, Neumann, C.M.T, Desdorf, L.M, Schioett, B, Salvino, J.M, Mortensen, O.V, Nissen, P, Shahsavar, A. | | Deposit date: | 2024-03-27 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure of the dopamine transporter with a novel atypical non-competitive inhibitor bound to the orthosteric site.
J.Neurochem., 2024
|
|
5HQO
 
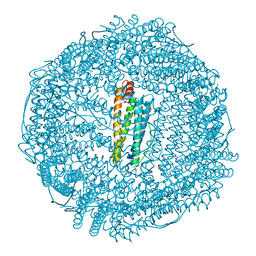 | | Crystal structure of IrCp*/I-Pd(allyl)-apo-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Maity, B, Fukumori, K, Abe, S, Ueno, T. | | Deposit date: | 2016-01-21 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Immobilization of two organometallic complexes into a single cage to construct protein-based microcompartments
Chem.Commun.(Camb.), 52, 2016
|
|
4U7T
 
 | | Crystal structure of DNMT3A-DNMT3L in complex with histone H3 | | Descriptor: | DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Guo, X, Wang, L, Yin, X, Li, J, Xiao, J, He, S, Wang, J, Xu, Y. | | Deposit date: | 2014-07-31 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insight into autoinhibition and histone H3-induced activation of DNMT3A
Nature, 517, 2015
|
|
5HQX
 
 | | Crystal structure of maize cytokinin oxidase/dehydrogenase 4 (ZmCKO4) in complex with phenylurea inhibitor HETDZ | | Descriptor: | 1-[2-(2-hydroxyethyl)phenyl]-3-(1,2,3-thiadiazol-5-yl)urea, Cytokinin dehydrogenase 4, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kopecny, D, Koncitikova, R, Briozzo, P. | | Deposit date: | 2016-01-22 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Novel thidiazuron-derived inhibitors of cytokinin oxidase/dehydrogenase.
Plant Mol.Biol., 92, 2016
|
|
2R8Q
 
 | | Structure of LmjPDEB1 in complex with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, Class I phosphodiesterase PDEB1, MAGNESIUM ION, ... | | Authors: | Wang, H, Yan, Z, Geng, J, Kunz, S, Seebeck, T, Ke, H. | | Deposit date: | 2007-09-11 | | Release date: | 2007-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the Leishmania major phosphodiesterase LmjPDEB1 and insight into the design of the parasite-selective inhibitors.
Mol.Microbiol., 66, 2007
|
|
5HLB
 
 | | E. coli PBP1b in complex with acyl-aztreonam and moenomycin | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, MOENOMYCIN, Penicillin-binding protein 1B | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2016-01-14 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Escherichia coli Penicillin-Binding Protein 1B: Structural Insights into Inhibition.
J. Biol. Chem., 2016
|
|
3LOW
 
 | |
4U5O
 
 | | IMPORTIN-ALPHA MINOR NLS SITE INHIBITOR | | Descriptor: | Importin subunit alpha-1, N~2~-[4-(pyridin-3-yl)benzyl]-D-lysinamide | | Authors: | Stewart, M, Valkov, E, Holvey, R.S. | | Deposit date: | 2014-07-25 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective Targeting of the TPX2 Site of Importin-alpha Using Fragment-Based Ligand Design.
Chemmedchem, 10, 2015
|
|
4U5R
 
 | |
2RLZ
 
 | | Solid-State MAS NMR structure of the dimer Crh | | Descriptor: | HPr-like protein crh | | Authors: | Loquet, A, Bardiaux, B, Gardiennet, C, Blanchet, C, Baldus, M, Nilges, M, Malliavin, T, Bockmann, A. | | Deposit date: | 2007-09-04 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-29 | | Method: | SOLID-STATE NMR | | Cite: | 3D structure determination of the Crh protein from highly ambiguous solid-state NMR restraints.
J.Am.Chem.Soc., 130, 2008
|
|
5HS7
 
 | |
5HLW
 
 | | Crystal structure of c-Met mutant Y1230H in complex with compound 14 | | Descriptor: | 1-[2-(1-ethylpiperidin-4-yl)ethyl]-3-(6-{[6-(thiophen-2-yl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]sulfanyl}-1,3-benzothiazol-2-yl)urea, CHLORIDE ION, Hepatocyte growth factor receptor | | Authors: | Vallee, F, Pouzieux, S, Marquette, J.P, Houtmann, J. | | Deposit date: | 2016-01-15 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery and Pharmacokinetic and Pharmacological Properties of the Potent and Selective MET Kinase Inhibitor 1-{6-[6-(4-Fluorophenyl)-[1,2,4]triazolo[4,3-b]pyridazin-3-ylsulfanyl]benzothiazol-2-yl}-3-(2-morpholin-4-ylethyl)urea (SAR125844).
J.Med.Chem., 59, 2016
|
|
2RMF
 
 | | Human Urocortin 1 | | Descriptor: | Urocortin | | Authors: | Grace, C.R.R, Perrin, M.H, Cantle, J.P, Vale, W.W, Rivier, J.E, Riek, R. | | Deposit date: | 2007-10-16 | | Release date: | 2008-01-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Common and divergent structural features of a series of corticotropin releasing factor-related peptides
J.Am.Chem.Soc., 129, 2007
|
|
5HM2
 
 | |
9B37
 
 | | Open state of kainate receptor GluK2 in complex with agonist glutamate and positive allosteric modulator BPAM344 bound to one concanavalin A dimer. Composite map. | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nadezhdin, K.D, Gangwar, S.P, Sobolevsky, A.I. | | Deposit date: | 2024-03-18 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (6.66 Å) | | Cite: | Kainate receptor channel opening and gating mechanism.
Nature, 630, 2024
|
|
1EI8
 
 | |
2RO1
 
 | | NMR Solution Structures of Human KAP1 PHD finger-bromodomain | | Descriptor: | Transcription intermediary factor 1-beta, ZINC ION | | Authors: | Zeng, L, Yap, K.L, Ivanov, A.V, Wang, X, Mujtaba, S, Plotnikova, O, Rauscher, F.J. | | Deposit date: | 2008-03-04 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural insights into human KAP1 PHD finger-bromodomain and its role in gene silencing
Nat.Struct.Mol.Biol., 15, 2008
|
|
4U9O
 
 | | Crystal structure of NqrA from Vibrio cholerae | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Na(+)-translocating NADH-quinone reductase subunit A | | Authors: | Fritz, G. | | Deposit date: | 2014-08-06 | | Release date: | 2014-12-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the V. cholerae Na+-pumping NADH:quinone oxidoreductase.
Nature, 516, 2014
|
|
5HN5
 
 | | Crystal structure of beta-decarboxylating dehydrogenase (TK0280) from Thermococcus kodakarensis complexed with Mn and isocitrate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Homoisocitrate dehydrogenase, ISOCITRIC ACID, ... | | Authors: | Shimizu, T, Tomita, T, Nishiyama, M. | | Deposit date: | 2016-01-18 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure and function of an ancestral-type beta-decarboxylating dehydrogenase from Thermococcus kodakarensis
Biochem. J., 474, 2017
|
|
2RP5
 
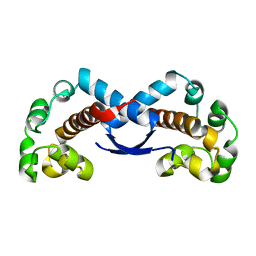 | |
9B36
 
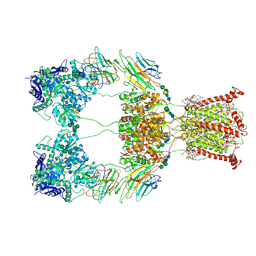 | | Open state of kainate receptor GluK2 in complex with agonist glutamate and positive allosteric modulator BPAM344 bound to two concanavalin A dimers. Composite map. | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nadezhdin, K.D, Gangwar, S.P, Sobolevsky, A.I. | | Deposit date: | 2024-03-18 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.29 Å) | | Cite: | Kainate receptor channel opening and gating mechanism.
Nature, 630, 2024
|
|
5HV9
 
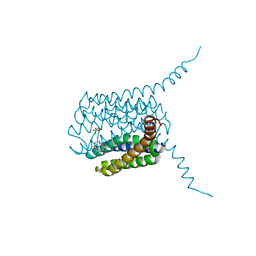 | | Human LTC4S mutant-S36E | | Descriptor: | GLUTATHIONE, Leukotriene C4 synthase, SULFATE ION | | Authors: | Thulasingam, M, Ahmad, H.R.S, Rinaldo-Matthis, A, Haeggstrom, J.Z. | | Deposit date: | 2016-01-28 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Phosphorylation of Leukotriene C4 Synthase at Serine 36 Impairs Catalytic Activity.
J.Biol.Chem., 291, 2016
|
|
