6I0M
 
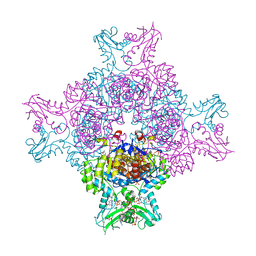 | | Structure of human IMP dehydrogenase, isoform 2, bound to GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, Inosine-5'-monophosphate dehydrogenase 2, ... | | Authors: | Buey, R.M, Fernandez-Justel, D, Revuelta, J.L. | | Deposit date: | 2018-10-26 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.567 Å) | | Cite: | A Nucleotide-Dependent Conformational Switch Controls the Polymerization of Human IMP Dehydrogenases to Modulate their Catalytic Activity.
J. Mol. Biol., 431, 2019
|
|
4L6U
 
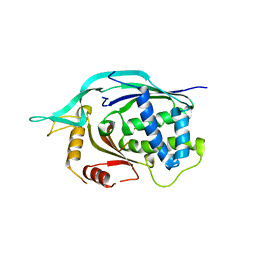 | | Crystal structure of AF1868: Cmr1 subunit of the Cmr RNA silencing complex | | Descriptor: | Putative uncharacterized protein | | Authors: | Sun, J, Jeon, J.H, Shin, M, Shin, H.C, Oh, B.H, Kim, J.S. | | Deposit date: | 2013-06-12 | | Release date: | 2014-02-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and CRISPR RNA-binding site of the Cmr1 subunit of the Cmr interference complex
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1VDX
 
 | |
6I0O
 
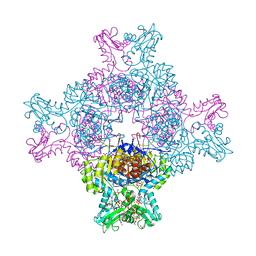 | | Structure of human IMP dehydrogenase, isoform 2, bound to GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase 2, SULFATE ION | | Authors: | Buey, R.M, Fernandez-Justel, D, Revuelta, J.L. | | Deposit date: | 2018-10-26 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.623 Å) | | Cite: | A Nucleotide-Dependent Conformational Switch Controls the Polymerization of Human IMP Dehydrogenases to Modulate their Catalytic Activity.
J. Mol. Biol., 431, 2019
|
|
8UMW
 
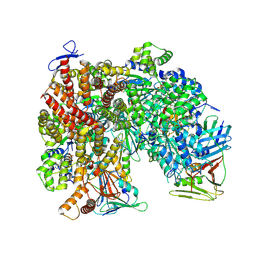 | | Atomic model of the human CTF18-RFC-PCNA-DNA ternary complex in the five-subunit binding state (state 4) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosome transmission fidelity protein 18 homolog, DNA (20-MER), ... | | Authors: | Wang, F, He, Q, Li, H. | | Deposit date: | 2023-10-18 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Cryo-EM reveals a nearly complete PCNA loading process and unique features of the human alternative clamp loader CTF18-RFC.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
2HB5
 
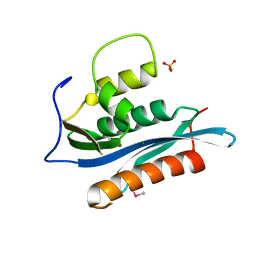 | | Crystal Structure of the Moloney Murine Leukemia Virus RNase H Domain | | Descriptor: | MAGNESIUM ION, Reverse transcriptase/ribonuclease H, SULFATE ION | | Authors: | Lim, D, Gregorio, G.G, Bingman, C.A, Martinez-Hackert, E, Hendrickson, W.A, Goff, S.P. | | Deposit date: | 2006-06-13 | | Release date: | 2006-08-29 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal Structure of the Moloney Murine Leukemia Virus RNase H Domain.
J.Virol., 80, 2006
|
|
2W4Y
 
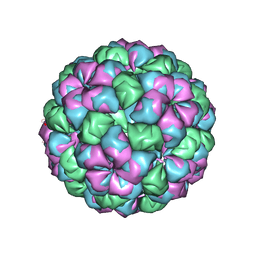 | | Caulobacter bacteriophage 5 - virus-like particle | | Descriptor: | ADENOSINE MONOPHOSPHATE, CALCIUM ION, CAULOBACTER 5 VIRUS-LIKE PARTICLE | | Authors: | Plevka, P, Kazaks, A, Dishlers, A, Liljas, L, Tars, K. | | Deposit date: | 2008-12-02 | | Release date: | 2009-07-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure of Bacteriophage Phicb5 Reveals a Role of the RNA Genome and Metal Ions in Particle Stability and Assembly.
J.Mol.Biol., 391, 2009
|
|
1NTQ
 
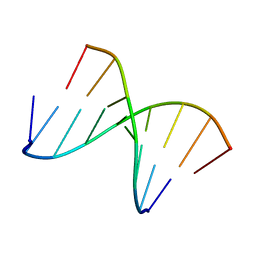 | | 5'(dCCUCCUU)3':3'(rAGGAGGAAA)5' | | Descriptor: | 5'-D(*CP*CP*UP*CP*CP*UP*U)-3', 5'-R(*AP*AP*AP*GP*GP*AP*GP*GP*A)-3' | | Authors: | Znosko, B.M, Barnes III, T.W, Krugh, T.R, Turner, D.H. | | Deposit date: | 2003-01-30 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Studies of DNA Single Strands and DNA:RNA Hybrids With and Without 1-Propynylation at C5 of Oligopyrimidines
J.Am.Chem.Soc., 125, 2003
|
|
433D
 
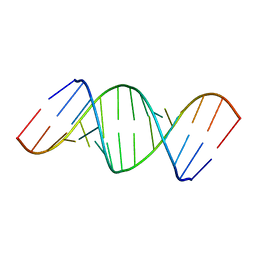 | |
2XMA
 
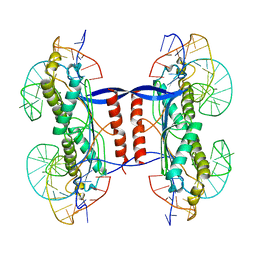 | | DEINOCOCCUS RADIODURANS ISDRA2 TRANSPOSASE RIGHT END DNA COMPLEX | | Descriptor: | DRA2 TRANSPOSASE RIGHT END RECOGNITION SITE, MAGNESIUM ION, TRANSPOSASE | | Authors: | Hickman, A.B, James, J.A, Barabas, O, Pasternak, C, Ton-Hoang, B, Chandler, M, Sommer, S, Dyda, F. | | Deposit date: | 2010-07-26 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA Recognition and the Precleavage State During Single-Stranded DNA Transposition in D. Radiodurans.
Embo J., 29, 2010
|
|
8PW8
 
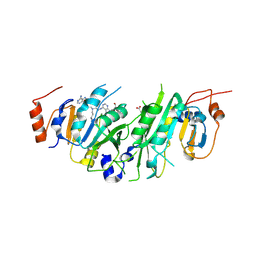 | | Crystal structure of the human METTL3-METTL14 in complex with a bisubstrate analogue (BA2) | | Descriptor: | (2~{S})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[2-[[9-[(2~{R},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]purin-6-yl]amino]ethyl]amino]-2-azanyl-butanoic acid, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Etheve-Quelquejeu, M, Caflisch, A. | | Deposit date: | 2023-07-19 | | Release date: | 2023-11-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The catalytic mechanism of the RNA methyltransferase METTL3.
Elife, 12, 2024
|
|
8PWA
 
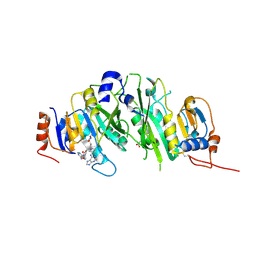 | | Crystal structure of the human METTL3-METTL14 in complex with a bisubstrate analogue (BA4) | | Descriptor: | (2~{S})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[3-[[9-[(2~{R},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-7~{H}-purin-6-yl]amino]propyl]amino]-2-azanyl-butanoic acid, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Bedi, R.K, Etheve-Quelquejeu, M, Caflisch, A. | | Deposit date: | 2023-07-19 | | Release date: | 2023-11-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The catalytic mechanism of the RNA methyltransferase METTL3.
Elife, 12, 2024
|
|
8PWB
 
 | | Crystal structure of the human METTL3-METTL14 in complex with a bisubstrate analogue (BA6) | | Descriptor: | (2~{S})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(7~{H}-purin-6-ylcarbamoyl)amino]-2-azanyl-butanoic acid, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Etheve-Quelquejeu, M, Caflisch, A. | | Deposit date: | 2023-07-19 | | Release date: | 2023-11-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The catalytic mechanism of the RNA methyltransferase METTL3.
Elife, 12, 2024
|
|
8PW9
 
 | | Crystal structure of the human METTL3-METTL14 in complex with a bisubstrate analogue (BA1) | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[[2-[[9-[(2~{R},3~{R},4~{S},5~{S})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]purin-6-yl]amino]ethylamino]methyl]oxolane-3,4-diol, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Bedi, R.K, Etheve-Quelquejeu, M, Caflisch, A. | | Deposit date: | 2023-07-19 | | Release date: | 2023-11-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The catalytic mechanism of the RNA methyltransferase METTL3.
Elife, 12, 2024
|
|
3ZL9
 
 | | Crystal structure of the nucleocapsid protein from Schmallenberg virus | | Descriptor: | NUCLEOCAPSID PROTEIN | | Authors: | Ariza, A, Tanner, S.J, Walter, C.T, Dent, K.C, Shepherd, D.A, Wu, W, Matthews, S.V, Hiscox, J.A, Green, T.J, Luo, M, Elliot, R.M, Ashcroft, A.E, Stonehouse, N.J, Ranson, N.A, Barr, J.N, Edwards, T.A. | | Deposit date: | 2013-01-29 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Nucleocapsid Protein Structures from Orthobunyaviruses Reveal Insight Into Ribonucleoprotein Architecture and RNA Polymerization.
Nucleic Acids Res., 41, 2013
|
|
3GKU
 
 | | Crystal structure of a probable RNA-binding protein from Clostridium symbiosum ATCC 14940 | | Descriptor: | Probable RNA-binding protein | | Authors: | Tan, K, Keigher, L, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-11 | | Release date: | 2009-03-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The crystal structure of a probable RNA-binding protein from Clostridium symbiosum ATCC 14940
To be Published
|
|
2JSG
 
 | | NMR solution structure of the anticodon of E.coli TRNA-VAL3 with 1 modification (M6A37) | | Descriptor: | 5'-R(*CP*CP*UP*CP*CP*CP*UP*UP*AP*CP*(6MZ)P*AP*GP*GP*AP*GP*G)-3' | | Authors: | Vendeix, F.A.P, Dziergowska, A, Gustilo, E.M, Graham, W.D, Sproat, B, Malkiewicz, A, Agris, P.F. | | Deposit date: | 2007-07-04 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Wobble-Position Modifications Pre-structure tRNA's Anticodon for Ribosome-Mediated Codon Binding
To be Published
|
|
2GIO
 
 | |
1J1H
 
 | | Solution structure of a tmRNA-binding protein, SmpB, from Thermus thermophilus | | Descriptor: | Small Protein B | | Authors: | Someya, T, Nameki, N, Hosoi, H, Suzuki, S, Hatanaka, H, Fujii, M, Terada, T, Shirouzu, M, Inoue, Y, Shibata, T, Kuramitsu, S, Yokoyama, S, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-12-04 | | Release date: | 2003-02-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a tmRNA-binding protein, SmpB, from Thermus thermophilus
FEBS Lett., 535, 2003
|
|
1XV6
 
 | |
2GIP
 
 | |
3IF0
 
 | |
2E48
 
 | | Crystal Structure of Human D-Amino Acid Oxidase: Substrate-Free Holoenzyme | | Descriptor: | D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kawazoe, T, Tsuge, H, Imagawa, T, Fukui, K. | | Deposit date: | 2006-12-05 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of d-DOPA oxidation by d-amino acid oxidase: Alternative pathway for dopamine biosynthesis.
Biochem.Biophys.Res.Commun., 355, 2007
|
|
2E4A
 
 | | Crystal Structure of Human D-Amino Acid Oxidase in complex with o-aminobenzoate | | Descriptor: | 2-AMINOBENZOIC ACID, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kawazoe, T, Tsuge, H, Imagawa, T, Fukui, K. | | Deposit date: | 2006-12-05 | | Release date: | 2007-03-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of d-DOPA oxidation by d-amino acid oxidase: Alternative pathway for dopamine biosynthesis.
Biochem.Biophys.Res.Commun., 355, 2007
|
|
2E82
 
 | | Crystal structure of human D-amino acid oxidase complexed with imino-DOPA | | Descriptor: | (2E)-3-(3,4-DIHYDROXYPHENYL)-2-IMINOPROPANOIC ACID, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kawazoe, T, Tsuge, H, Imagawa, T, Kuramitsu, S, Fukui, K. | | Deposit date: | 2007-01-16 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of d-DOPA oxidation by d-amino acid oxidase: Alternative pathway for dopamine biosynthesis.
Biochem.Biophys.Res.Commun., 355, 2007
|
|
