5OI1
 
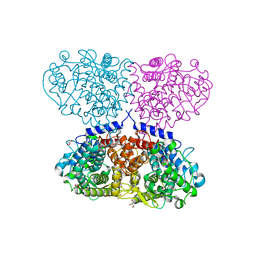 | |
4W9U
 
 | |
4ZXI
 
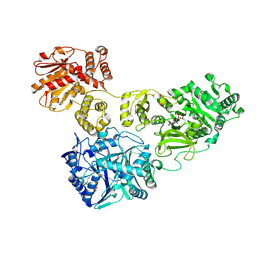 | | Crystal Structure of holo-AB3403 a four domain nonribosomal peptide synthetase bound to AMP and Glycine | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, 4'-PHOSPHOPANTETHEINE, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Drake, E.J, Miller, B.R, Allen, C.L, Gulick, A.M. | | Deposit date: | 2015-05-20 | | Release date: | 2015-12-30 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of two distinct conformations of holo-non-ribosomal peptide synthetases.
Nature, 529, 2016
|
|
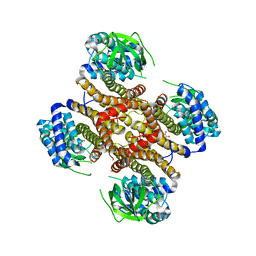
5O9J
 
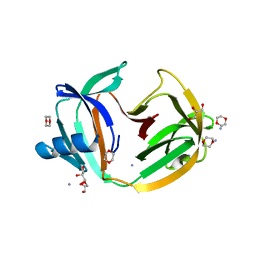 | | Crystal structure of transcription factor IIB Mja mini-intein | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, AMMONIUM ION, ... | | Authors: | Mikula, K.M, Iwai, H, Zhou, D, Wlodawer, A. | | Deposit date: | 2017-06-19 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Persistence of Homing Endonucleases in Transcription Factor IIB Inteins.
J. Mol. Biol., 429, 2017
|
|
4ZY8
 
 | |
5O9O
 
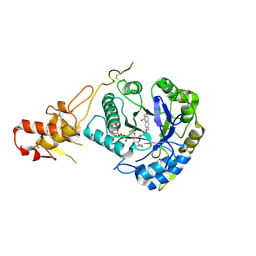 | | Crystal structure of ScGas2 in complex with compound 7. | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-[4-(naphthalen-2-ylmethoxymethyl)-1,2,3-triazol-1-yl]-3,5-bis(oxidanyl)oxan-4-yl]oxy-3,5-bis(oxidanyl)oxan-4-yl]oxy-oxane-3,4,5-triol, 1,2-ETHANEDIOL, 1,3-beta-glucanosyltransferase GAS2 | | Authors: | Delso, I, Valero-Gonzalez, J, Fang, W, Gomollon-Bel, F, Castro-Lopez, J, Navratilova, I, van Aalten, D, Tejero, T, Merino, P, Hurtado-Guerrero, R. | | Deposit date: | 2017-06-19 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibitors against Fungal Cell Wall Remodeling Enzymes.
ChemMedChem, 13, 2018
|
|
5WJ5
 
 | |
2BUG
 
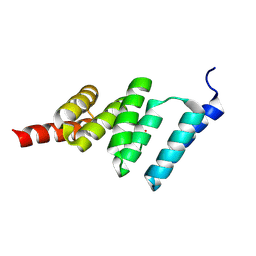 | | Solution structure of the TPR domain from Protein phosphatase 5 in complex with Hsp90 derived peptide | | Descriptor: | HSP90, SERINE/THREONINE PROTEIN PHOSPHATASE 5 | | Authors: | Cliff, M.J, Harris, R, Barford, D, Ladbury, J.E, Williams, M.A. | | Deposit date: | 2005-06-13 | | Release date: | 2006-03-16 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Conformational Diversity in the Tpr Domain-Mediated Interaction of Protein Phosphatase 5 with Hsp90.
Structure, 14, 2006
|
|
4ZYH
 
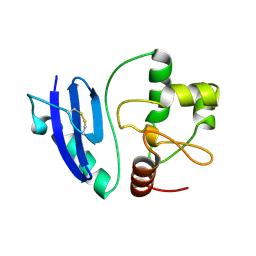 | |
5A9K
 
 | | Structural basis for DNA strand separation by a hexameric replicative helicase | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, REPLICATION PROTEIN E1 | | Authors: | Chaban, Y, Stead, J.A, Ryzhenkova, K, Whelan, F, Lamber, K, Antson, F, Sanders, C.M, Orlova, E.V. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (19 Å) | | Cite: | Structural Basis for DNA Strand Separation by a Hexameric Replicative Helicase.
Nucleic Acids Res., 43, 2015
|
|
6PCT
 
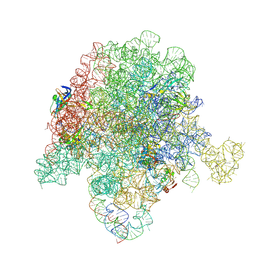 | | E. coli 50S ribosome bound to compound 41q | | Descriptor: | (2S)-2-[(3S,4R,5E,10E,12E,14S,26aR)-14-hydroxy-4,12-dimethyl-1,7,16,22-tetraoxo-4,7,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,3H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosin-3-yl]propyl isoquinolin-3-ylcarbamate, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2019-06-18 | | Release date: | 2020-06-17 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
5O9Y
 
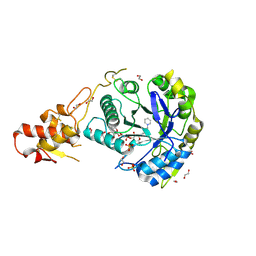 | | Crystal structure of ScGas2 in complex with compound 11 | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-3,5-bis(oxidanyl)-6-[4-(3-pyridin-1-ium-1-ylpropyl)-1,2,3-triazol-1-yl]oxan-4-yl]oxy-3,5-bis(oxidanyl)oxan-4-yl]oxy-oxane-3,4,5-triol, 1,2-ETHANEDIOL, 1,3-beta-glucanosyltransferase GAS2, ... | | Authors: | Delso, I, Valero-Gonzalez, J, Gomollon-Bel, F, Castro-Lopez, J, Fang, W, Navratilova, I, Van Aalten, D, Tejero, T, Merino, P, Hurtado-Guerrero, R. | | Deposit date: | 2017-06-20 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Inhibitors against Fungal Cell Wall Remodeling Enzymes.
ChemMedChem, 13, 2018
|
|
5OKP
 
 | |
5OKT
 
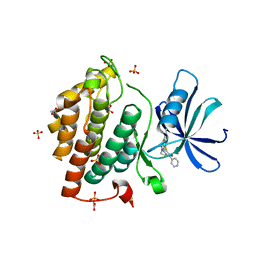 | | Crystal structure of human Casein Kinase I delta in complex with IWP-2 | | Descriptor: | ACETATE ION, Casein kinase I isoform delta, GLYCEROL, ... | | Authors: | Pichlo, C, Brunstein, E, Baumann, U. | | Deposit date: | 2017-07-25 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Discovery of Inhibitor of Wnt Production 2 (IWP-2) and Related Compounds As Selective ATP-Competitive Inhibitors of Casein Kinase 1 (CK1) delta / epsilon.
J. Med. Chem., 61, 2018
|
|
5OAB
 
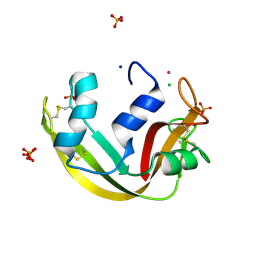 | | A novel crystal form of human RNase6 at atomic resolution | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Prats-Ejarque, G, Moussaoui, M, Boix, E. | | Deposit date: | 2017-06-21 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.111 Å) | | Cite: | Characterization of an RNase with two catalytic centers. Human RNase6 catalytic and phosphate-binding site arrangement favors the endonuclease cleavage of polymeric substrates.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
5OLQ
 
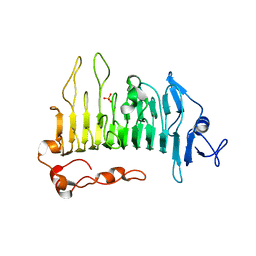 | | Rhamnogalacturonan lyase | | Descriptor: | CALCIUM ION, PHOSPHATE ION, Rhamnogalacturonan lyase | | Authors: | Basle, A, Luis, A.S, Gilbert, H.J. | | Deposit date: | 2017-07-28 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides.
Nat Microbiol, 3, 2018
|
|
5ACA
 
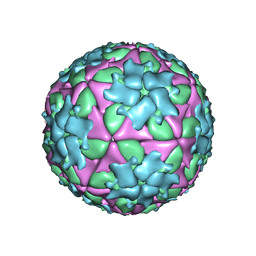 | | Structure-based energetics of protein interfaces guide Foot-and-Mouth disease virus vaccine design | | Descriptor: | VP1, VP2, VP3, ... | | Authors: | Kotecha, A, Seago, J, Scott, K, Burman, A, Loureiro, S, Ren, J, Porta, C, Ginn, H.M, Jackson, T, Perez-Martin, E, Siebert, C.A, Paul, G, Huiskonen, J.T, Jones, I.M, Esnouf, R.M, Fry, E.E, Maree, F.F, Charleston, B, Stuart, D.I. | | Deposit date: | 2015-08-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure-Based Energetics of Protein Interfaces Guide Foot-and-Mouth Disease Vaccine Design
Nat.Struct.Mol.Biol., 22, 2015
|
|
5NTJ
 
 | |
5WQK
 
 | |
4UOX
 
 | |
4UHH
 
 | | Structural studies of a thermophilic esterase from Thermogutta terrifontis (cacodylate complex) | | Descriptor: | CACODYLIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sayer, C, Isupov, M.N, Bonch-Osmolovskaya, E, Littlechild, J.A. | | Deposit date: | 2015-03-24 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Structural Studies of a Thermophilic Esterase from a New Planctomycetes Species, Thermogutta Terrifontis.
FEBS J., 282, 2015
|
|
5WRG
 
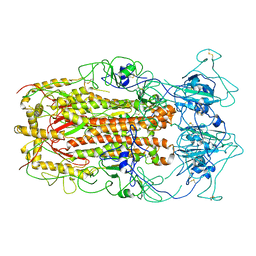 | | SARS-CoV spike glycoprotein | | Descriptor: | Spike glycoprotein | | Authors: | Gui, M, Song, W, Xiang, Y, Wang, X. | | Deposit date: | 2016-12-01 | | Release date: | 2017-01-11 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding.
Cell Res., 27, 2017
|
|
5NUH
 
 | |
5OB8
 
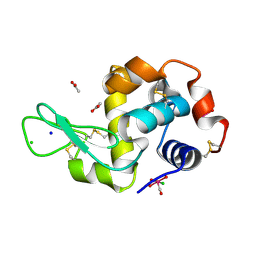 | | X-ray structure of the adduct formed upon reaction of lysozyme with the compound fac-[RuII(CO)3Cl2(N3-MIM), MIM=methyl-imidazole (crystals grown using NaCl) | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Pontillo, N, Ferraro, G, Merlino, A. | | Deposit date: | 2017-06-26 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Ru-Based CO releasing molecules with azole ligands: interaction with proteins and the CO release mechanism disclosed by X-ray crystallography.
Dalton Trans, 46, 2017
|
|
4UIM
 
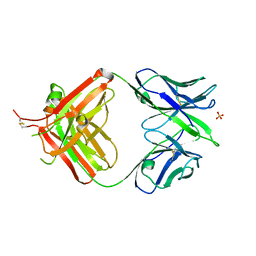 | | crystal structure of quinine-dependent Fab 314.3 | | Descriptor: | FAB 314.3, SULFATE ION | | Authors: | Zhu, J, Zhu, J, Bougie, D.W, Aster, R.H, Springer, T.A. | | Deposit date: | 2015-03-30 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Quinine-Dependent Antibody Binding to Platelet Integrin Alphaiib Beta3
Blood, 126, 2015
|
|
