3BV8
 
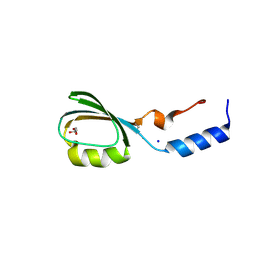 | | Crystal structure of the N-terminal domain of tetrahydrodipicolinate acetyltransferase from Staphylococcus aureus | | Descriptor: | GLYCEROL, SODIUM ION, Tetrahydrodipicolinate acetyltransferase | | Authors: | Cuff, M.E, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the N-terminal domain of tetrahydrodipicolinate acetyltransferase from Staphylococcus aureus.
TO BE PUBLISHED
|
|
3C7H
 
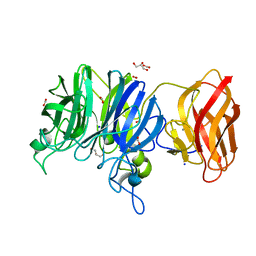 | | Crystal structure of glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from Bacillus subtilis in complex with AXOS-4-0.5. | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase, FORMIC ACID, ... | | Authors: | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | Deposit date: | 2008-02-07 | | Release date: | 2008-11-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
8XPE
 
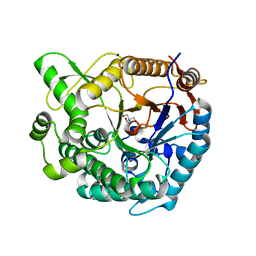 | | Crystal structure of Tris-bound TsaBgl (DATA III) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SODIUM ION, beta-glucosidase | | Authors: | Nam, K.H. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of Tris binding in beta-glucosidases.
Biochem.Biophys.Res.Commun., 700, 2024
|
|
8XPC
 
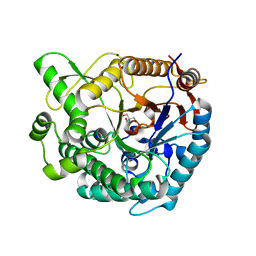 | | Crystal structure of Tris-bound TsaBgl (DATA I) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SODIUM ION, beta-glucosidase | | Authors: | Nam, K.H. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analysis of Tris binding in beta-glucosidases.
Biochem.Biophys.Res.Commun., 700, 2024
|
|
8XPD
 
 | | Crystal structure of Tris-bound TsaBgl (DATA II) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SODIUM ION, beta-glucosidase | | Authors: | Nam, K.H. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of Tris binding in beta-glucosidases.
Biochem.Biophys.Res.Commun., 700, 2024
|
|
8YBG
 
 | |
2ZHI
 
 | |
8YBH
 
 | |
3BIB
 
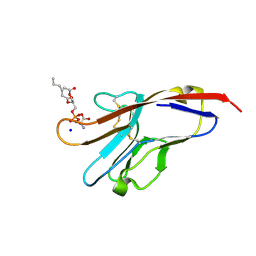 | | Tim-4 in complex with phosphatidylserine | | Descriptor: | 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, SODIUM ION, T-cell immunoglobulin and mucin domain-containing protein 4 | | Authors: | Santiago, C, Ballesteros, A, Kaplan, G.G, Freeman, G.J, Casasnovas, J.M. | | Deposit date: | 2007-11-30 | | Release date: | 2008-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of T Cell Immunoglobulin Mucin Protein 4 Show a Metal-Ion-Dependent Ligand Binding Site where Phosphatidylserine Binds.
Immunity, 27, 2007
|
|
3B5G
 
 | |
3B6E
 
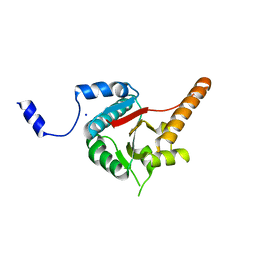 | | Crystal structure of human DECH-box RNA Helicase MDA5 (Melanoma differentiation-associated protein 5), DECH-domain | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, SODIUM ION | | Authors: | Karlberg, T, Welin, M, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Human DECH-box RNA Helicase MDA5 (Melanoma differentiation-associated protein 5), DECH-domain.
To be Published
|
|
5FTT
 
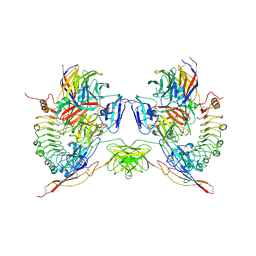 | | Octameric complex of Latrophilin 3 (Lec, Olf) , Unc5D (Ig, Ig2, TSP1) and FLRT2 (LRR) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADHESION G PROTEIN-COUPLED RECEPTOR L3, CALCIUM ION, ... | | Authors: | Jackson, V.A, Mehmood, S, Chavent, M, Roversi, P, Carrasquero, M, del Toro, D, Seyit-Bremer, G, Ranaivoson, F.M, Comoletti, D, Sansom, M.S.P, Robinson, C.V, Klein, R, Seiradake, E. | | Deposit date: | 2016-01-15 | | Release date: | 2016-05-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Super-Complexes of Adhesion Gpcrs and Neural Guidance Receptors
Nat.Commun., 7, 2016
|
|
7D10
 
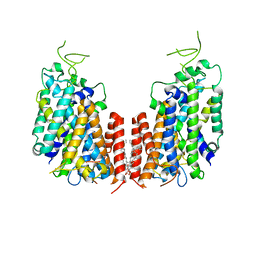 | | Human NKCC1 | | Descriptor: | PALMITIC ACID, Solute carrier family 12 member 2 | | Authors: | Zhang, S, Yang, M. | | Deposit date: | 2020-09-12 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | The structural basis of function and regulation of neuronal cotransporters NKCC1 and KCC2.
Commun Biol, 4, 2021
|
|
3BKC
 
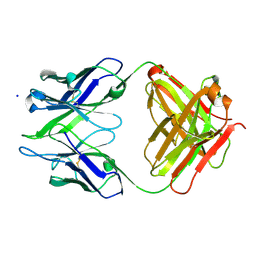 | | Crystal structure of anti-amyloid beta FAB WO2 (P21, FormB) | | Descriptor: | SODIUM ION, WO2 IgG2a Fab fragment Heavy Chain, WO2 IgG2a Fab fragment Light Chain Kappa | | Authors: | Miles, L.A, Wun, K.S, Crespi, G.A, Fodero-Tavoletti, M, Galatis, D, Bageley, C.J, Beyreuther, K, Masters, C.L, Cappai, R, McKinstry, W.J, Barnham, K.J, Parker, M.W. | | Deposit date: | 2007-12-06 | | Release date: | 2008-04-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Amyloid-beta-anti-amyloid-beta complex structure reveals an extended conformation in the immunodominant B-cell epitope.
J.Mol.Biol., 377, 2008
|
|
7D18
 
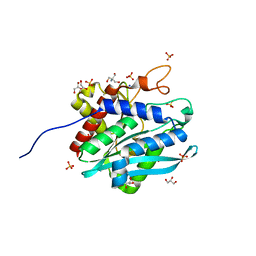 | | Crystal structure of Acidobacteriales bacterium glutaminyl cyclase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Peptidase M28, ... | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.332 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
3BN1
 
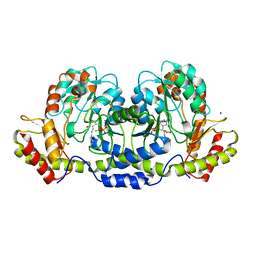 | | Crystal structure of GDP-perosamine synthase | | Descriptor: | 2-OXOGLUTARIC ACID, ACETATE ION, Perosamine synthetase, ... | | Authors: | Cook, P.D, Holden, H.M. | | Deposit date: | 2007-12-13 | | Release date: | 2008-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | GDP-Perosamine Synthase: Structural Analysis and Production of a Novel Trideoxysugar
Biochemistry, 47, 2008
|
|
7D12
 
 | | NMR solution structures of CAG RNA-DB213 binding complex | | Descriptor: | N'-{(Z)-amino[4-(amino{[3-(dimethylammonio)propyl]iminio}methyl)phenyl]methylidene}-N,N-dimethylpropane-1,3-diaminium, RNA (5'-R(*GP*CP*AP*GP*CP*AP*GP*CP*UP*UP*CP*GP*GP*CP*AP*GP*CP*AP*GP*C)-3'), SODIUM ION | | Authors: | Chan, H.Y.E, Guo, P. | | Deposit date: | 2020-09-12 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | CAG RNAs induce DNA damage and apoptosis by silencing NUDT16 expression in polyglutamine degeneration.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DPD
 
 | | Human MCM9 N-terminal domain | | Descriptor: | DNA helicase MCM9, SODIUM ION, ZINC ION | | Authors: | Li, J, Liu, L, Liu, Y. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural study of the N-terminal domain of human MCM8/9 complex.
Structure, 29, 2021
|
|
3C6V
 
 | | Crystal structure of AU4130/APC7354, a probable enzyme from the thermophilic fungus Aspergillus fumigatus | | Descriptor: | CHLORIDE ION, Probable tautomerase/dehalogenase AU4130, SODIUM ION, ... | | Authors: | Singer, A.U, Binkowski, T.A, Skarina, T, Kagan, O, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-05 | | Release date: | 2008-02-19 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of AU4130/APC7354, a probable enzyme from the thermophilic fungus Aspergillus fumigatus.
To be Published
|
|
3BOS
 
 | |
7DJJ
 
 | | Structure of four truncated and mutated forms of quenching protein lumenal domains | | Descriptor: | Protein SUPPRESSOR OF QUENCHING 1, chloroplastic, SODIUM ION, ... | | Authors: | Yu, G.M, Pan, X.W, Li, M. | | Deposit date: | 2020-11-20 | | Release date: | 2022-06-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.69806433 Å) | | Cite: | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
7DJM
 
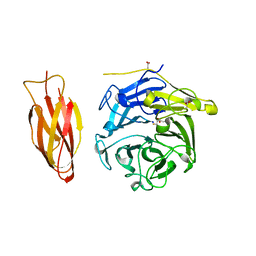 | | Structure of four truncated and mutated forms of quenching protein | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACETATE ION, Protein SUPPRESSOR OF QUENCHING 1, ... | | Authors: | Yu, G.M, Pan, X.W, Li, M. | | Deposit date: | 2020-11-20 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.70000112 Å) | | Cite: | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
7DJK
 
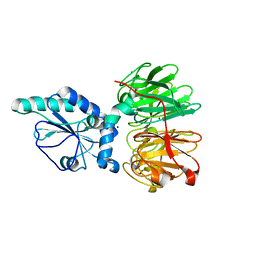 | | Structure of four truncated and mutated forms of quenching protein | | Descriptor: | CHLORIDE ION, Protein SUPPRESSOR OF QUENCHING 1, chloroplastic, ... | | Authors: | Yu, G.M, Pan, X.W, Li, M. | | Deposit date: | 2020-11-20 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.80145121 Å) | | Cite: | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
7DJL
 
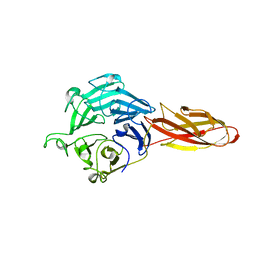 | | Structure of four truncated and mutated forms of quenching protein | | Descriptor: | CHLORIDE ION, Protein SUPPRESSOR OF QUENCHING 1, chloroplastic, ... | | Authors: | Yu, G.M, Pan, X.W, Li, M. | | Deposit date: | 2020-11-20 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.96077824 Å) | | Cite: | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
7DTF
 
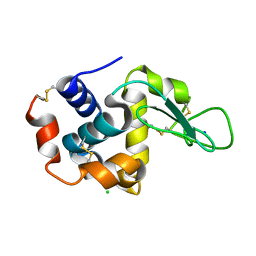 | |
