8VHU
 
 | |
7CRE
 
 | |
7CY3
 
 | |
7CY9
 
 | |
8WFU
 
 | |
8WFW
 
 | |
8WFT
 
 | |
5M0M
 
 | | Structure-based evolution of a hybrid steroid series of Autotaxin inhibitors | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, CALCIUM ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Keune, W.-J, Heidebrecht, T, Perrakis, A. | | Deposit date: | 2016-10-05 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rational Design of Autotaxin Inhibitors by Structural Evolution of Endogenous Modulators.
J. Med. Chem., 60, 2017
|
|
8WFV
 
 | |
5M0S
 
 | | Structure-based evolution of a hybrid steroid series of Autotaxin inhibitors | | Descriptor: | CALCIUM ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, GLYCEROL, ... | | Authors: | Keune, W.-J, Heidebrecht, T, Perrakis, A. | | Deposit date: | 2016-10-05 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rational Design of Autotaxin Inhibitors by Structural Evolution of Endogenous Modulators.
J. Med. Chem., 60, 2017
|
|
5LSH
 
 | | human lysozyme in complex with a tetrasaccharide fragment of the O-chain of LPS from Klebsiella pneumoniae | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Zhang, R, Nifantiev, N.E, Krylov, V, Luetteke, T, Scheidig, A.J, Siebert, H.-C. | | Deposit date: | 2016-08-26 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.061 Å) | | Cite: | Lysozyme's lectin-like characteristics facilitates its immune defense function.
Q. Rev. Biophys., 50, 2017
|
|
7D2A
 
 | | CBM32 of AlyQ in complex with 4,5-unsaturated mannuronic acid | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid, AlyQ, CALCIUM ION, ... | | Authors: | Teh, A.H, Sim, P.F. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural basis for binding uronic acids by family 32 carbohydrate-binding modules.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
2YMS
 
 | |
7CY1
 
 | | Crystal Structure of MglC from Myxococcus xanthus | | Descriptor: | Mutual gliding motility protein C, SODIUM ION | | Authors: | Thakur, K.G, Kapoor, S, Kodesia, A. | | Deposit date: | 2020-09-03 | | Release date: | 2021-01-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural characterization of Myxococcus xanthus MglC, a component of the polarity control system, and its interactions with its paralog MglB.
J.Biol.Chem., 2021
|
|
6H0M
 
 | | 17beta-hydroxysteroid dehydrogenase type 14 mutant K158A in complex with Nicotinamide Adenine Dinucleotide | | Descriptor: | 17-beta-hydroxysteroid dehydrogenase 14, DIMETHYL SULFOXIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Badran, M, Klebe, G, Heine, A, Marchais-Oberwinkler, S. | | Deposit date: | 2018-07-10 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | 17beta-hydroxysteroid dehydrogenase type 14 mutant K158A in complex with Nicotinamide Adenine Dinucleotide
To Be Published
|
|
5EYQ
 
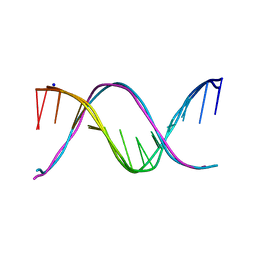 | | Racemic crystal structures of Pribnow box consensus promoter sequence (Pnna) | | Descriptor: | Complementary strand, Pribnow box template strand, SODIUM ION | | Authors: | Mandal, P.K, Collie, G.W, Kauffmann, B, Srivastava, S.C, Huc, I. | | Deposit date: | 2015-11-25 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure elucidation of the Pribnow box consensus promoter sequence by racemic DNA crystallography.
Nucleic Acids Res., 44, 2016
|
|
7D3F
 
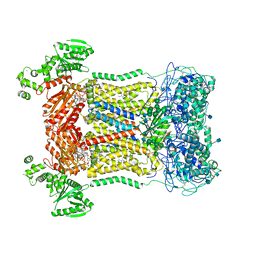 | | Cryo-EM structure of human DUOX1-DUOXA1 in high-calcium state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Dual oxidase 1, ... | | Authors: | Chen, L, Wu, J.X. | | Deposit date: | 2020-09-19 | | Release date: | 2020-12-09 | | Last modified: | 2021-06-23 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structures of human dual oxidase 1 complex in low-calcium and high-calcium states.
Nat Commun, 12, 2021
|
|
7D79
 
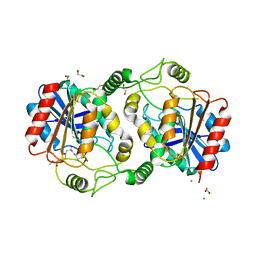 | | The structure of DcsB complex with its substrate analogue | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DltD domain-containing protein, ... | | Authors: | Tang, Y, Zhou, J.H, Wang, G.Q. | | Deposit date: | 2020-10-03 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.10411429 Å) | | Cite: | A Polyketide Cyclase That Forms Medium-Ring Lactones.
J.Am.Chem.Soc., 143, 2021
|
|
7DTB
 
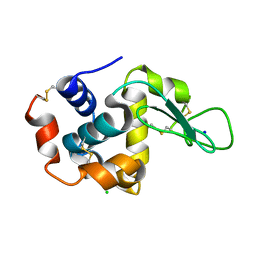 | |
8VR2
 
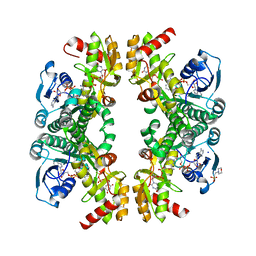 | | Crystal structure of the Pcryo_0617 oxidoreductase/decarboxylase from Psychrobacter cryohalolentis K5 in the presence of NAD and UDP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NAD-dependent epimerase/dehydratase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Bockhaus, N.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2024-01-20 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical Investigation of the Enzymes Required for the Production of 2,3,4-triacetoamido-2,3,4-trideoxy-l-arabinose in Psychrobacter cryohalolentis K5
To Be Published
|
|
8VR3
 
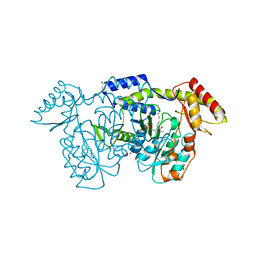 | | crystal structure of the Pcryo_0618 aminotransferase from Psychrobacter cryohalolentis K5 in the presence of its internal aldimine | | Descriptor: | 1,2-ETHANEDIOL, 2,2'-(1,4-diazepane-1,4-diyl)di(ethane-1-sulfonic acid), CHLORIDE ION, ... | | Authors: | Bockhaus, N.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2024-01-20 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical Investigation of the Enzymes Required for the Production of 2,3,4-triacetoamido-2,3,4-trideoxy-l-arabinose in Psychrobacter cryohalolentis K5
To Be Published
|
|
5MIG
 
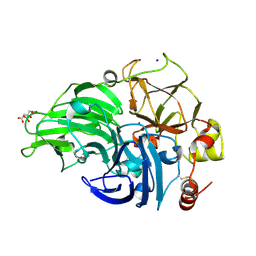 | | The study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi.The 16-th structure of the series with total exposition time 453 min. The crystal was quick refreezing before this data collection. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Polyakov, K.M, Gavryushov, S, Fedorova, T.V, Glazunova, O.A, Popov, A.N. | | Deposit date: | 2016-11-28 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural study of the X-ray-induced enzymatic reduction of molecular oxygen to water by Steccherinum murashkinskyi laccase: insights into the reaction mechanism.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
8VR5
 
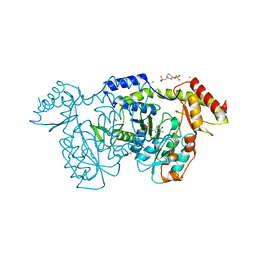 | | crystal structure of the Pcryo_0618 aminotransferase from Psychrobacter cryohalolentis K5 in the presence of PMP and glutamate | | Descriptor: | 1,2-ETHANEDIOL, 2,2'-(1,4-diazepane-1,4-diyl)di(ethane-1-sulfonic acid), 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Bockhaus, N.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2024-01-20 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical Investigation of the Enzymes Required for the Production of 2,3,4-triacetoamido-2,3,4-trideoxy-l-arabinose in Psychrobacter cryohalolentis K5
To Be Published
|
|
5GLN
 
 | | Crystal structure of CoXyl43, GH43 beta-xylosidase/alpha-arabinofuranosidase from a compostmicrobial metagenome in complex with xylotriose, calcium-bound form | | Descriptor: | ACETATE ION, CALCIUM ION, Glycoside hydrolase family 43, ... | | Authors: | Matsuzawa, T, Kishine, N, Fujimoto, Z, Yaoi, K. | | Deposit date: | 2016-07-12 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of metagenomic beta-xylosidase/ alpha-l-arabinofuranosidase activated by calcium.
J. Biochem., 162, 2017
|
|
5GLP
 
 | | Crystal structure of CoXyl43, GH43 beta-xylosidase/alpha-arabinofuranosidase from a compostmicrobial metagenome in complex with l-arabinose, calcium-bound form | | Descriptor: | ACETATE ION, CALCIUM ION, Glycoside hydrolase family 43, ... | | Authors: | Matsuzawa, T, Kishine, N, Fujimoto, Z, Yaoi, K. | | Deposit date: | 2016-07-12 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of metagenomic beta-xylosidase/ alpha-l-arabinofuranosidase activated by calcium.
J. Biochem., 162, 2017
|
|
