5C37
 
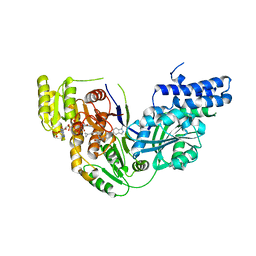 | | Structure of the beta-ketoacyl reductase domain of human fatty acid synthase bound to a spiro-imidazolone inhibitor | | Descriptor: | 6-{[(3R)-1-(cyclopropylcarbonyl)pyrrolidin-3-yl]methyl}-5-[4-(1-methyl-1H-indazol-5-yl)phenyl]-4,6-diazaspiro[2.4]hept-4-en-7-one, CHLORIDE ION, Fatty acid synthase, ... | | Authors: | Schubert, C, Milligan, C.M, Vo, K, Grasberger, B. | | Deposit date: | 2015-06-17 | | Release date: | 2016-06-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and synthesis of a series of bioavailable fatty acid synthase (FASN) KR domain inhibitors for cancer therapy.
Bioorg.Med.Chem.Lett., 28, 2018
|
|
3ZDG
 
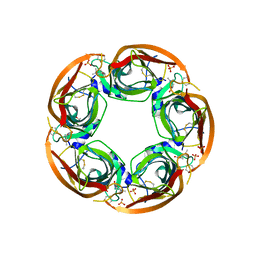 | | Crystal Structure of Ls-AChBP complexed with carbamoylcholine analogue 3-(dimethylamino)butyl dimethylcarbamate (DMABC) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(dimethylamino)butyl dimethylcarbamate, ACETYLCHOLINE BINDING PROTEIN, ... | | Authors: | Ussing, C.A, Hansen, C.P, Petersen, J.G, Jensen, A.A, Rohde, L.A.H, Ahring, P.K, Nielsen, E.O, Kastrup, J.S, Gajhede, M, Frolund, B, Balle, T. | | Deposit date: | 2012-11-26 | | Release date: | 2013-02-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Synthesis, Pharmacology, and Biostructural Characterization of Novel Alpha(4)Beta(2) Nicotinic Acetylcholine Receptor Agonists.
J.Med.Chem., 56, 2013
|
|
3ZDH
 
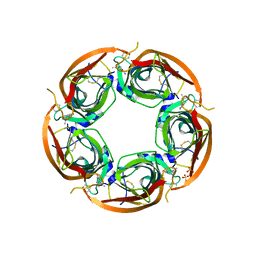 | | Crystal structure of Ls-AChBP complexed with carbamoylcholine analogue N,N-dimethyl-4-(1-methyl-1H-imidazol-2-yloxy)butan-2-amine | | Descriptor: | (2R)-N,N-dimethyl-4-(1-methylimidazol-2-yl)oxy-butan-2-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE BINDING PROTEIN, ... | | Authors: | Ussing, C.A, Hansen, C.P, Petersen, J.G, Jensen, A.A, Rohde, L.A.H, Ahring, P.K, Nielsen, E.O, Kastrup, J.S, Gajhede, M, Frolund, B, Balle, T. | | Deposit date: | 2012-11-26 | | Release date: | 2013-02-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Synthesis, Pharmacology, and Biostructural Characterization of Novel Alpha(4)Beta(2) Nicotinic Acetylcholine Receptor Agonists.
J.Med.Chem., 56, 2013
|
|
3CY2
 
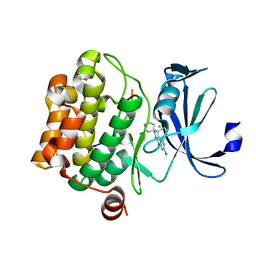 | | Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and a beta carboline ligand II | | Descriptor: | (4R)-7-chloro-9-methyl-1-oxo-1,2,4,9-tetrahydrospiro[beta-carboline-3,4'-piperidine]-4-carbonitrile, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Filippakopoulos, P, Bullock, A, Fedorov, O, Huber, K, Bracher, F, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-25 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | 7,8-Dichloro-1-oxo-beta-carbolines as a Versatile Scaffold for the Development of Potent and Selective Kinase Inhibitors with Unusual Binding Modes
J.Med.Chem., 55, 2012
|
|
6W9I
 
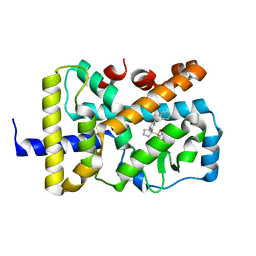 | |
6W9H
 
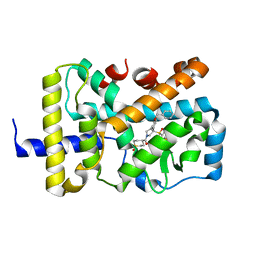 | | SUBSTITUTED BENZYLOXYTRICYCLIC COMPOUNDS AS RETINOIC ACID-RELATED ORPHAN RECEPTOR GAMMA T AGONISTS | | Descriptor: | 4-{(3R)-3-[4-(benzyloxy)phenyl]-3-[(4-fluorophenyl)sulfonyl]pyrrolidine-1-carbonyl}-1lambda~6~-thiane-1,1-dione, Nuclear receptor ROR-gamma, Steroid receptor coactivator 1 fusion | | Authors: | Sack, J.S. | | Deposit date: | 2020-03-23 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Substituted benzyloxytricyclic compounds as retinoic acid-related orphan receptor gamma t (ROR gamma t) agonists.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
3E7C
 
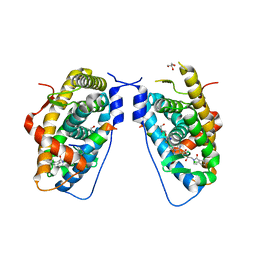 | | Glucocorticoid Receptor LBD bound to GSK866 | | Descriptor: | 5-amino-N-[(2S)-2-({[(2,6-dichlorophenyl)carbonyl](ethyl)amino}methyl)-3,3,3-trifluoro-2-hydroxypropyl]-1-(4-fluorophenyl)-1H-pyrazole-4-carboxamide, GLYCEROL, Glucocorticoid receptor, ... | | Authors: | Madauss, K.P, Williams, S.P, Mclay, I, Stewart, E.L, Bledsoe, R.K. | | Deposit date: | 2008-08-18 | | Release date: | 2008-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The first X-ray crystal structure of the glucocorticoid receptor bound to a non-steroidal agonist.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4P8O
 
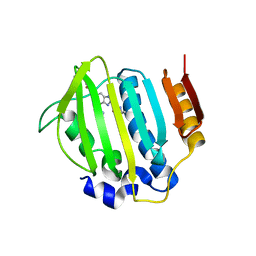 | | S. aureus gyrase bound to an aminobenzimidazole urea inhibitor | | Descriptor: | 1-ethyl-3-[5-(5-fluoropyridin-3-yl)-7-(pyrimidin-2-yl)-1H-benzimidazol-2-yl]urea, DNA gyrase subunit B | | Authors: | Jacobs, M.D. | | Deposit date: | 2014-03-31 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Second-generation antibacterial benzimidazole ureas: discovery of a preclinical candidate with reduced metabolic liability.
J.Med.Chem., 57, 2014
|
|
7TEK
 
 | |
7TEL
 
 | |
6VL3
 
 | | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ000986436 | | Descriptor: | 5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-6-oxo-2-[2-(trifluoromethyl)phenyl]-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Jayaraman, S, Rankovic, Z, White, S.W. | | Deposit date: | 2020-01-22 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
6VJH
 
 | | The crystal structure of the 2009 H1N1/California PA endonuclease wild type in complex with SJ000986192 | | Descriptor: | 2-[2-[(cyclohexylmethyl-$l^{3}-oxidanyl)carbonylamino]propan-2-yl]-~{N}-[2-(5-methoxy-4-oxidanyl-cyclohexa-1,3,5-trien-1-yl)ethyl]-5-oxidanyl-6-oxidanylidene-pyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2020-01-16 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
6VIV
 
 | | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ000986192 | | Descriptor: | 2-[2-[(cyclohexylmethyl-$l^{3}-oxidanyl)carbonylamino]propan-2-yl]-~{N}-[2-(5-methoxy-4-oxidanyl-cyclohexa-1,3,5-trien-1-yl)ethyl]-5-oxidanyl-6-oxidanylidene-pyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2020-01-14 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
6WYR
 
 | | Crystal structure of anti-Muscle Specific Kinase (MuSK) Fab, MuSK1A | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MuSK1A heavy chain, ... | | Authors: | Vieni, C, Ekiert, D. | | Deposit date: | 2020-05-13 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Affinity maturation is required for pathogenic monovalent IgG4 autoantibody development in myasthenia gravis.
J.Exp.Med., 217, 2020
|
|
4HHY
 
 | | Crystal structure of PARP catalytic domain in complex with novel inhibitors | | Descriptor: | (9aR)-1-[(1-{2-fluoro-5-[(4-oxo-3,4-dihydrophthalazin-1-yl)methyl]benzoyl}piperidin-4-yl)carbonyl]-1,2,3,8,9,9a-hexahydro-7H-benzo[de][1,7]naphthyridin-7-one, DI(HYDROXYETHYL)ETHER, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Liu, Q.F, Chen, T.T, Xu, Y.C. | | Deposit date: | 2012-10-10 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3637 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of a Series of Benzo[de][1,7]naphthyridin-7(8H)-ones Bearing a Functionalized Longer Chain Appendage as Novel PARP1 Inhibitors.
J.Med.Chem., 56, 2013
|
|
4HHZ
 
 | | Crystal structure of PARP catalytic domain in complex with novel inhibitors | | Descriptor: | N-{(2S)-1-[4-(4-fluorophenyl)-3,6-dihydropyridin-1(2H)-yl]-1-oxopropan-2-yl}-2-[(9aR)-7-oxo-2,3,7,8,9,9a-hexahydro-1H-benzo[de][1,7]naphthyridin-1-yl]acetamide, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Liu, Q.F, Chen, T.T, Xu, Y.C. | | Deposit date: | 2012-10-10 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7199 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of a Series of Benzo[de][1,7]naphthyridin-7(8H)-ones Bearing a Functionalized Longer Chain Appendage as Novel PARP1 Inhibitors.
J.Med.Chem., 56, 2013
|
|
4HWS
 
 | |
6WEP
 
 | | Crystal structure of TS-DHFR from Cryptosporidium hominis with Apo-TS site | | Descriptor: | Bifunctional dihydrofolate reductase-thymidylate synthase, METHOTREXATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Czyzyk, D.J, Ruiz, V.G, Kumar, V.P, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Targeting the TS dimer interface in bifunctional Cryptosporidium hominis TS-DHFR from parasitic protozoa: Virtual screening identifies novel TS allosteric inhibitor
Bioorg.Med.Chem.Lett., 30, 2020
|
|
7U5Z
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with JLJ353 | | Descriptor: | 2-chloro-4-({5-[(2,6-difluorophenyl)methyl]-1,3-oxazol-2-yl}amino)benzonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Hollander, K, Carter, Z, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2022-03-03 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, synthesis, and biological testing of biphenylmethyloxazole inhibitors targeting HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 84, 2023
|
|
6WYT
 
 | |
4XY8
 
 | | Crystal Structure of the bromodomain of BRD9 in complex with a 2-amine-9H-purine ligand | | Descriptor: | 6-(5-bromo-2-methoxyphenyl)-9H-purin-2-amine, BROMIDE ION, Bromodomain-containing protein 9 | | Authors: | Picaud, S, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 9H-Purine Scaffold Reveals Induced-Fit Pocket Plasticity of the BRD9 Bromodomain.
J.Med.Chem., 58, 2015
|
|
6WIJ
 
 | | The crystal structure of the 2009/H1N1/California PA endonuclease mutant I38T in complex with SJ000986448 | | Descriptor: | 5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-6-oxo-2-[2-(trifluoromethyl)phenyl]-3,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2020-04-10 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
6WJ4
 
 | | The crystal structure of the 2009/H1N1/California PA endonuclease wild type in complex with SJ000986448 | | Descriptor: | 5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-6-oxo-2-[2-(trifluoromethyl)phenyl]-3,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2020-04-11 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
4QG7
 
 | |
4QGA
 
 | |
