8W4S
 
 | | Crystal structure of PDE5A in complex with CVT-313 | | Descriptor: | 2,2'-{[6-{[(4-methoxyphenyl)methyl]amino}-9-(propan-2-yl)-9H-purin-2-yl]azanediyl}di(ethan-1-ol), MAGNESIUM ION, ZINC ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Drug repurposing and structure-based discovery of new PDE4 and PDE5 inhibitors.
Eur.J.Med.Chem., 262, 2023
|
|
8W4T
 
 | | Crystal structure of PDE5A in complex with a novel inhibitor | | Descriptor: | 2-[bis(2-hydroxyethyl)amino]-6-[(4-methoxyphenyl)methylamino]-9-propan-2-yl-7~{H}-purin-8-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Drug repurposing and structure-based discovery of new PDE4 and PDE5 inhibitors.
Eur.J.Med.Chem., 262, 2023
|
|
6I0L
 
 | | Crystal structure of human carbonic anhydrase I in complex with the 1-[4-chloro-3-(trifluoromethyl)phenyl]-3-[2-(4-sulfamoylphenyl)ethyl]urea inhibitor | | Descriptor: | 1-[4-chloranyl-3-(trifluoromethyl)phenyl]-3-[2-(4-sulfamoylphenyl)ethyl]urea, ACETATE ION, Carbonic anhydrase 1, ... | | Authors: | Ferraroni, M, Supuran, C.T, Bozdag, M, Chiapponi, D. | | Deposit date: | 2018-10-26 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Carbonic anhydrase inhibitors based on sorafenib scaffold: Design, synthesis, crystallographic investigation and effects on primary breast cancer cells.
Eur.J.Med.Chem., 182, 2019
|
|
4Z0K
 
 | |
2H04
 
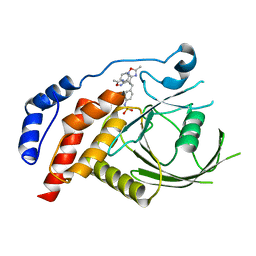 | | Structural studies of protein tyrosine phosphatase beta catalytic domain in complex with inhibitors | | Descriptor: | Protein tyrosine phosphatase, receptor type, B,, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M, Gray, J.L, Peters, K.G, Maier, M.B, Amarasinghe, K.D, Clark, C.M, Nichols, R. | | Deposit date: | 2006-05-13 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and synthesis of potent, non-peptidic inhibitors of HPTPbeta.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4XIP
 
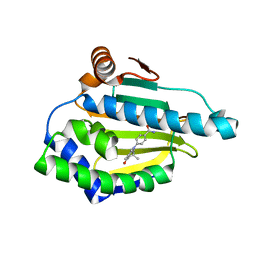 | | Discovery of novel oxazepine and diazepine carboxamides as two new classes of heat shock protein 90 inhibitors | | Descriptor: | 4-(3,6,6-trimethyl-4-oxo-4,5,6,7-tetrahydro-1H-indol-1-yl)benzamide, Heat shock protein HSP 90-alpha | | Authors: | Zuccola, H.J, Neubert, T. | | Deposit date: | 2015-01-07 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of novel oxazepine and diazepine carboxamides as two new classes of heat shock protein 90 inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4ZAE
 
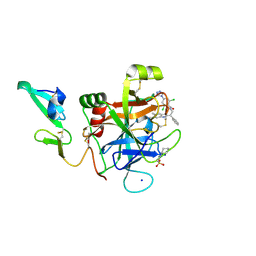 | | Development of a novel class of potent and selective FIXa inhibitors | | Descriptor: | 2,6-dichloro-N-[(2R)-2-(5,6-dimethyl-1H-benzimidazol-2-yl)-2-phenylethyl]-4-(4H-1,2,4-triazol-4-yl)benzamide, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Coagulation factor IX, ... | | Authors: | Hruza, A, Reichert, P. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Development of a novel class of potent and selective FIXa inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
2FVD
 
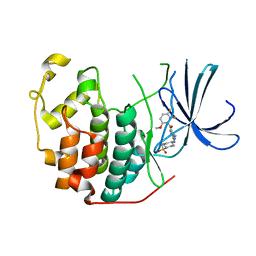 | | Cyclin Dependent Kinase 2 (CDK2) with diaminopyrimidine inhibitor | | Descriptor: | (4-AMINO-2-{[1-(METHYLSULFONYL)PIPERIDIN-4-YL]AMINO}PYRIMIDIN-5-YL)(2,3-DIFLUORO-6-METHOXYPHENYL)METHANONE, Cell division protein kinase 2 | | Authors: | Crowther, R.L, Lukacs, C.M, Kammlott, R.U. | | Deposit date: | 2006-01-30 | | Release date: | 2006-10-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of [4-Amino-2-(1-methanesulfonylpiperidin-4-ylamino)pyrimidin-5-yl](2,3-difluoro-6- methoxyphenyl)methanone (R547), a potent and selective cyclin-dependent kinase inhibitor with significant in vivo antitumor activity.
J.Med.Chem., 49, 2006
|
|
4U01
 
 | | HCV NS3/4A serine protease in complex with 6570 | | Descriptor: | (2S,3aS,10Z,11aS,12aR)-2-({8-fluoro-7-methoxy-2-[4-(propan-2-yl)-1,3-thiazol-2-yl]quinolin-4-yl}oxy)-5-methyl-N-[(1-methylcyclopropyl)sulfonyl]-4,14-dioxo-1,2,3,3a,4,5,6,7,8,9,11a,12,13,14-tetradecahydro-12aH-cyclopropa[m]pyrrolo[1,2-c][1,3,6]triazacyclotetradecine-12a-carboxamide, CHLORIDE ION, NS4A protein, ... | | Authors: | Parsy, C.C, Alexandre, F.-R, Brandt, G, Caillet, C, Chaves, D, Derock, M, Gloux, D, Griffon, Y, Lallos, L.B, Leroy, F, Liuzzi, M, Loi, A.-G, Mayes, B, Moulat, L, Moussa, A, Chiara, M, Roques, V, Rosinovsky, E, Seifer, M, Stewart, A, Wang, J, Standring, D, Surleraux, D. | | Deposit date: | 2014-07-11 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and structural diversity of the hepatitis C virus NS3/4A serine protease inhibitor series leading to clinical candidate IDX320.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
3U8W
 
 | | Crystal Structure of p38a Mitogen-Activated Protein Kinase in Complex with a Triazolopyridazinone inhibitor | | Descriptor: | 3-[3-(2-chloro-6-fluorophenyl)-5-ethyl-6-oxo-5,6-dihydro[1,2,4]triazolo[4,3-b]pyridazin-7-yl]-N-cyclopropyl-4-methylbenzamide, Mitogen-activated protein kinase 14 | | Authors: | Mohr, C, Jordan, S. | | Deposit date: | 2011-10-17 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification of triazolopyridazinones as potent p38alpha inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3VV6
 
 | | Crystal Structure of beta secetase in complex with 2-amino-3-methyl-6-((1S, 2R)-2-phenylcyclopropyl)pyrimidin-4(3H)-one | | Descriptor: | 2-amino-3-methyl-6-[(1S,2R)-2-phenylcyclopropyl]pyrimidin-4(3H)-one, Beta-secretase 1, GLYCEROL, ... | | Authors: | Yonezawa, S, Yamamoto, T, Yamakawa, H, Muto, C, Hosono, M, Hattori, K, Higashino, K, Sakagami, M, Togame, H, Tanaka, Y, Nakano, T, Takemoto, H, Arisawa, M, Shuto, S. | | Deposit date: | 2012-07-17 | | Release date: | 2012-10-24 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformational restriction approach to beta-secretase (BACE1) inhibitors: effect of a cyclopropane ring to induce an alternative binding mode
J.Med.Chem., 55, 2012
|
|
5AA8
 
 | | Structure of C1156Y,L1198F Mutant Human Anaplastic Lymphoma Kinase in Complex with PF-06463922 ((10R)-7-amino-12-fluoro-2,10,16-trimethyl- 15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo(4,3-h)(2,5,11) benzoxadiazacyclotetradecine-3-carbonitrile). | | Descriptor: | (10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo[4,3-h][2,5,11]benzoxadiazacyclotetradecine-3-carbonitrile, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M, Deng, Y.-L, Liu, W, Brooun, A, Stewart, A. | | Deposit date: | 2015-07-23 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Resensitization to Crizotinib by the Lorlatinib Alk Resistance Mutation L1198F.
N.Engl.J.Med., 374, 2016
|
|
3VO3
 
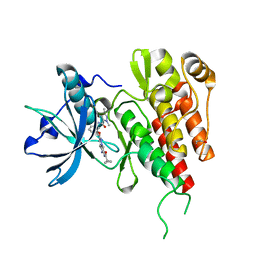 | | Crystal Structure of the Kinase domain of Human VEGFR2 with imidazo[1,2-b]pyridazine derivative | | Descriptor: | 1,2-ETHANEDIOL, N-[3-({2-[(cyclopropylcarbonyl)amino]imidazo[1,2-b]pyridazin-6-yl}oxy)phenyl]-1,3-dimethyl-1H-pyrazole-5-carboxamide, Vascular endothelial growth factor receptor 2 | | Authors: | Oki, H, Okada, K. | | Deposit date: | 2012-01-19 | | Release date: | 2013-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of N-[5-({2-[(cyclopropylcarbonyl)amino]imidazo[1,2-b]pyridazin-6-yl}oxy)-2-methylphenyl]-1,3-dimethyl-1H-pyrazole-5-carboxamide (TAK-593), a highly potent VEGFR2 kinase inhibitor
Bioorg.Med.Chem., 21, 2013
|
|
3VAP
 
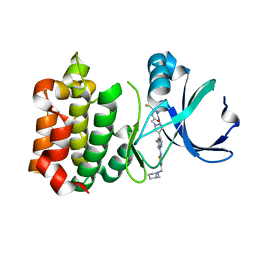 | | Synthesis and SAR Studies of imidazo-[1,2-a]-pyrazine Aurora kinase inhibitors with improved off target kinase selectivity | | Descriptor: | 3-(1-{2-[(3-fluoropyridinium-4-yl)amino]-2-oxoethyl}-1H-pyrazol-4-yl)-6-methyl-8-[(3-{[(1R,3R)-3-methylpiperidinium-1-yl]methyl}-1,2-thiazol-5-yl)amino]imidazo[1,2-a]pyrazin-1-ium, Aurora kinase A | | Authors: | Voss, M.E, Rainka, M.P, Fleming, M, Peterson, L.H, Belanger, D.B, Siddiqui, M.A, Hruza, A, Voigt, J, Basso, A.D, Gray, K. | | Deposit date: | 2011-12-29 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Synthesis and SAR studies of imidazo-[1,2-a]-pyrazine Aurora kinase inhibitors with improved off-target kinase selectivity.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3VV8
 
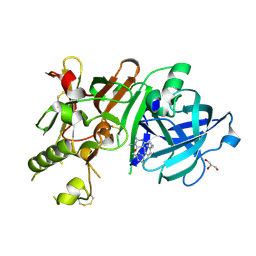 | | Crystal structure of beta secetase in complex with 2-amino-3-methyl-6-((1S,2R)-2-(3'-methylbiphenyl-4-yl)cyclopropyl)pyrimidin-4(3H)-one | | Descriptor: | 2-amino-3-methyl-6-[(1S,2R)-2-(3'-methylbiphenyl-4-yl)cyclopropyl]pyrimidin-4(3H)-one, Beta-secretase 1, GLYCEROL | | Authors: | Yonezawa, S, Yamamoto, T, Yamakawa, H, Muto, C, Hosono, M, Hattori, K, Higashino, K, Sakagami, M, Togame, H, Tanaka, Y, Nakano, T, Takemoto, H, Arisawa, M, Shuto, S. | | Deposit date: | 2012-07-17 | | Release date: | 2012-10-24 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational restriction approach to beta-secretase (BACE1) inhibitors: effect of a cyclopropane ring to induce an alternative binding mode.
J.Med.Chem., 55, 2012
|
|
5AR2
 
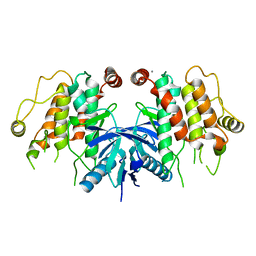 | | RIP2 Kinase Catalytic Domain (1 - 310) | | Descriptor: | CALCIUM ION, RECEPTOR-INTERACTING SERINE/THREONINE-PROTEIN KINASE 2 | | Authors: | Charnley, A.K, Convery, M.A, Lakdawala Shah, A, Jones, E, Hardwicke, P, Bridges, A, Votta, B.J, Gough, P.J, Marquis, R.W, Bertin, J, Casillas, L. | | Deposit date: | 2015-09-23 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal Structures of Human Rip2 Kinase Catalytic Domain Complexed with ATP-Competitive Inhibitors: Foundations for Understanding Inhibitor Selectivity.
Bioorg.Med.Chem., 23, 2015
|
|
5AR7
 
 | | RIP2 Kinase Catalytic Domain (1 - 310) complex with Biaryl Urea | | Descriptor: | 1-(5-TERT-BUTYL-1,2-OXAZOL-3-YL)-3-(4-PYRIDIN-4-YLOXYPHENYL)UREA, RECEPTOR-INTERACTING SERINE/THREONINE-PROTEIN KINASE 2 | | Authors: | Charnley, A.K, Convery, M.A, Lakdawala Shah, A, Jones, E, Hardwicke, P, Bridges, A, Votta, B.J, Gough, P.J, Marquis, R.W, Bertin, J, Casillas, L. | | Deposit date: | 2015-09-24 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal Structures of Human Rip2 Kinase Catalytic Domain Complexed with ATP-Competitive Inhibitors: Foundations for Understanding Inhibitor Selectivity.
Bioorg.Med.Chem., 23, 2015
|
|
3VV7
 
 | | Crystal Structure of beta secetase in complex with 2-amino-6-((1S,2R)-2-(3'-methoxybiphenyl-3-yl)cyclopropyl)-3-methylpyrimidin-4(3H)-one | | Descriptor: | 2-amino-6-[(1S,2R)-2-(3'-methoxybiphenyl-3-yl)cyclopropyl]-3-methylpyrimidin-4(3H)-one, Beta-secretase 1, GLYCEROL, ... | | Authors: | Yonezawa, S, Yamamoto, T, Yamakawa, H, Muto, C, Hosono, M, Hattori, K, Higashino, K, Sakagami, M, Togame, H, Tanaka, Y, Nakano, T, Takemoto, H, Arisawa, M, Shuto, S. | | Deposit date: | 2012-07-17 | | Release date: | 2012-10-24 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational restriction approach to beta-secretase (BACE1) inhibitors: effect of a cyclopropane ring to induce an alternative binding mode.
J.Med.Chem., 55, 2012
|
|
7KU4
 
 | |
7KWK
 
 | |
3WKE
 
 | | Crystal structure of soluble epoxide hydrolase in complex with t-AUCB | | Descriptor: | 4-[(trans-4-{[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]dec-1-ylcarbamoyl]amino}cyclohexyl)oxy]benzoic acid, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Amano, Y, Yamaguchi, T, Tanabe, E. | | Deposit date: | 2013-10-18 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural insights into binding of inhibitors to soluble epoxide hydrolase gained by fragment screening and X-ray crystallography.
Bioorg.Med.Chem., 22, 2014
|
|
3WV3
 
 | | Crystal structure of the catalytic domain of MMP-13 complexed with N-(3-methoxybenzyl)-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidine-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Collagenase 3, ... | | Authors: | Oki, H, Tanaka, Y. | | Deposit date: | 2014-05-12 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Thieno[2,3-d]pyrimidine-2-carboxamides bearing a carboxybenzene group at 5-position: highly potent, selective, and orally available MMP-13 inhibitors interacting with the S1′′ binding site.
Bioorg.Med.Chem., 22, 2014
|
|
7U8W
 
 | | hTRAP1 with inhibitors | | Descriptor: | Heat shock protein 75 kDa, mitochondrial, [5-(4-fluoro-2H-isoindole-2-carbonyl)-2-hydroxyphenyl](5-fluoro-2H-isoindol-2-yl)methanone | | Authors: | Deng, J, Matts, R, Peng, S. | | Deposit date: | 2022-03-09 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.706 Å) | | Cite: | Elucidation of novel TRAP1-Selective inhibitors that regulate mitochondrial processes.
Eur.J.Med.Chem., 258, 2023
|
|
7U8X
 
 | | hTRAP1 with inhibitors | | Descriptor: | Heat shock protein 75 kDa, mitochondrial, [5-(4-fluoro-2H-isoindole-2-carbonyl)-2-hydroxyphenyl](5-methoxy-2H-isoindol-2-yl)methanone | | Authors: | Deng, J, Matts, R, Peng, S. | | Deposit date: | 2022-03-09 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Elucidation of novel TRAP1-Selective inhibitors that regulate mitochondrial processes.
Eur.J.Med.Chem., 258, 2023
|
|
7U8V
 
 | | hTRAP1 with inhibitors | | Descriptor: | (4-hydroxy-1,3-phenylene)bis[(2H-isoindol-2-yl)methanone], Heat shock protein 75 kDa, mitochondrial | | Authors: | Deng, J, Matts, R, Peng, S. | | Deposit date: | 2022-03-09 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.448 Å) | | Cite: | Elucidation of novel TRAP1-Selective inhibitors that regulate mitochondrial processes.
Eur.J.Med.Chem., 258, 2023
|
|
