1NQS
 
 | | Structural Characterisation of the Holliday Junction formed by the sequence d(TCGGTACCGA) at 1.97 A | | Descriptor: | 5'-d(TpCpGpGpTpApCpCpGpA)-3', CALCIUM ION | | Authors: | Cardin, C.J, Gale, B.C, Thorpe, J.H, Texieira, S.C.M, Gan, Y, Moraes, M.I.A.A, Brogden, A.L. | | Deposit date: | 2003-01-22 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Analysis of two Holliday junctions formed by the sequences TCGGTACCGA and CCGGTACCGG
To be Published
|
|
1EPL
 
 | | A STRUCTURAL COMPARISON OF 21 INHIBITOR COMPLEXES OF THE ASPARTIC PROTEINASE FROM ENDOTHIA PARASITICA | | Descriptor: | ENDOTHIAPEPSIN, PS1, PRO-LEU-GLU-PSA-ARG-LEU | | Authors: | Al-Karadaghi, S, Cooper, J.B, Strop, P, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural comparison of 21 inhibitor complexes of the aspartic proteinase from Endothia parasitica.
Protein Sci., 3, 1994
|
|
6VVO
 
 | | Structure of the human clamp loader (Replication Factor C, RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen, PCNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Stone, N.P, Kelch, B.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-02-26 | | Last modified: | 2020-03-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the human clamp loader bound to the sliding clamp: a further twist on AAA+ mechanism
Biorxiv, 2020
|
|
6CD0
 
 | | Crystal structure of Medicago truncatula serine hydroxymethyltransferase 3 (MtSHMT3), PLP-internal aldimine and apo form | | Descriptor: | ACETATE ION, FORMIC ACID, Serine hydroxymethyltransferase | | Authors: | Ruszkowski, M, Sekula, B, Ruszkowska, A, Dauter, Z. | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Chloroplastic Serine Hydroxymethyltransferase FromMedicago truncatula: A Structural Characterization.
Front Plant Sci, 9, 2018
|
|
5OHC
 
 | |
7ZLP
 
 | | Crystal structure of SOCS2:ElonginB:ElonginC in complex with compound 9 | | Descriptor: | Elongin-B, Elongin-C, PHOSPHATE ION, ... | | Authors: | Ramachandran, S, Ciulli, A, Makukhin, N. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure-based design of a phosphotyrosine-masked covalent ligand targeting the E3 ligase SOCS2.
Nat Commun, 14, 2023
|
|
5OIW
 
 | | Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) D182A variant in complex with glucosylglycerate | | Descriptor: | (2R)-2-(alpha-D-glucopyranosyloxy)-3-hydroxypropanoic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Cereija, T.B, Macedo-Ribeiro, S, Pereira, P.J.B. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The structural characterization of a glucosylglycerate hydrolase provides insights into the molecular mechanism of mycobacterial recovery from nitrogen starvation.
Iucrj, 6, 2019
|
|
4G2N
 
 | | Crystal structure of putative D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding from Polaromonas sp. JS6 66 | | Descriptor: | CHLORIDE ION, D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-07-12 | | Release date: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding from Polaromonas sp. JS6 66
To be Published
|
|
7B18
 
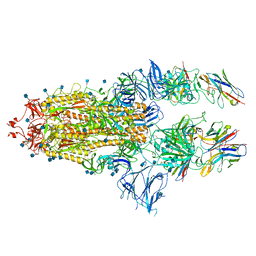 | | SARS-CoV-spike bound to two neutralising nanobodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody against SARS-CoV-2 VHH E, ... | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
1EQH
 
 | | THE 2.7 ANGSTROM MODEL OF OVINE COX-1 COMPLEXED WITH FLURBIPROFEN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLURBIPROFEN, PROSTAGLANDIN H2 SYNTHASE-1, ... | | Authors: | Loll, P.J, Selinsky, B.S, Gupta, K, Sharkey, C.T. | | Deposit date: | 2000-04-04 | | Release date: | 2001-04-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of NSAID binding by prostaglandin H2 synthase: time-dependent and time-independent inhibitors elicit identical enzyme conformations.
Biochemistry, 40, 2001
|
|
6W3U
 
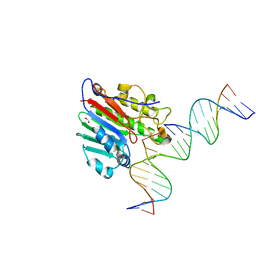 | | APE1 exonuclease substrate complex R237C | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Freudenthal, B.D, Whitaker, A.M. | | Deposit date: | 2020-03-09 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular and structural characterization of disease-associated APE1 polymorphisms.
DNA Repair (Amst.), 91-92, 2020
|
|
6P43
 
 | |
6W72
 
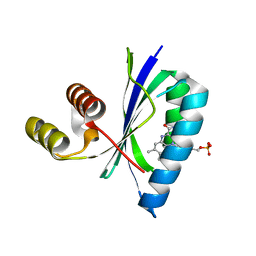 | | BlsA photo-activated state | | Descriptor: | BLUF domain-containing protein, FLAVIN MONONUCLEOTIDE | | Authors: | Chitrakar, I, French, J.B. | | Deposit date: | 2020-03-18 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Basis for the Regulation of Biofilm Formation and Iron Uptake in A. baumannii by the Blue-Light-Using Photoreceptor, BlsA.
Acs Infect Dis., 6, 2020
|
|
1NQ2
 
 | | Two RTH Mutants with Impaired Hormone Binding | | Descriptor: | ARSENIC, SODIUM ION, Thyroid hormone receptor beta-1, ... | | Authors: | Huber, B.R, Sandler, B, West, B.L, Cunha-Lima, S.T, Nguyen, H.T, Apriletti, J.W, Baxter, J.D, Fletterick, R.J. | | Deposit date: | 2003-01-21 | | Release date: | 2003-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Two resistance to thyroid hormone mutants with impaired hormone binding
Mol.Endocrinol., 17, 2003
|
|
1EOG
 
 | | CRYSTAL STRUCTURE OF PI CLASS GLUTATHIONE TRANSFERASE | | Descriptor: | GLUTATHIONE S-TRANSFERASE | | Authors: | Rossjohn, J, McKinstry, W.J, Oakley, A.J, Parker, M.W, Stenberg, G, Mannervik, B, Dragani, B, Cocco, R, Aceto, A. | | Deposit date: | 2000-03-22 | | Release date: | 2000-10-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of thermolabile mutants of human glutathione transferase P1-1.
J.Mol.Biol., 302, 2000
|
|
6W3D
 
 | | Rd1NTF2_05 with long sheet | | Descriptor: | Rd1NTF2_05 | | Authors: | Bick, M.J, Basanta, B, Sankaran, B, Baker, D. | | Deposit date: | 2020-03-09 | | Release date: | 2020-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | An enumerative algorithm for de novo design of proteins with diverse pocket structures.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7AIN
 
 | | Structure of Human Potassium Chloride Transporter KCC3 S45D/T940D/T997D in NaCl (Reference Map) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 2 of Solute carrier family 12 member 6, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chi, G, Man, H, Ebenhoch, R, Reggiano, G, Pike, A.C.W, Wang, D, McKinley, G, Mukhopadhyay, S.M.M, MacLean, E.M, Chalk, R, Moreau, C, Snee, M, Bohstedt, T, Singh, N.K, Abrusci, P, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Marsden, B.D, Burgess-Brown, N.A, DiMaio, F, Duerr, K.L, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-09-28 | | Release date: | 2021-06-02 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Phospho-regulation, nucleotide binding and ion access control in potassium-chloride cotransporters.
Embo J., 40, 2021
|
|
1NUG
 
 | |
2BO8
 
 | | DISSECTION OF MANNOSYLGLYCERATE SYNTHASE: AN ARCHETYPAL MANNOSYLTRANSFERASE | | Descriptor: | CHLORIDE ION, GUANOSINE 5'-(TRIHYDROGEN DIPHOSPHATE), P'-D-MANNOPYRANOSYL ESTER, ... | | Authors: | Flint, J, Taylor, E, Yang, M, Bolam, D.N, Tailford, L.E, Martinez-Fleites, C, Dodson, E.J, Davis, B.G, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2005-04-08 | | Release date: | 2005-06-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural dissection and high-throughput screening of mannosylglycerate synthase.
Nat. Struct. Mol. Biol., 12, 2005
|
|
1O0R
 
 | | Crystal structure of the catalytic domain of bovine beta1,4-galactosyltransferase complex with UDP-galactose | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GALACTOSE-URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Ramakrishnan, B, Balaji, P.V, Qasba, P.K. | | Deposit date: | 2003-02-24 | | Release date: | 2003-03-04 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF BETA 1,4-GALACTOSYLTRANSFERASE COMPLEX WITH UDP-GAL
REVEALS AN OLIGOSACCHARIDE ACCEPTOR BINDING SITE
J.Mol.Biol., 318, 2002
|
|
6CO7
 
 | | Structure of the nvTRPM2 channel in complex with Ca2+ | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zhang, Z, Toth, B, Szollosi, A, Chen, J, Csanady, L. | | Deposit date: | 2018-03-12 | | Release date: | 2018-05-16 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structure of a TRPM2 channel in complex with Ca2+explains unique gating regulation.
Elife, 7, 2018
|
|
6CI6
 
 | | Crystal structure of equine serum albumin in complex with nabumetone | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, Serum albumin, ... | | Authors: | Venkataramany, B.S, Czub, M.P, Shabalin, I.G, Handing, K.B, Steen, E.H, Cooper, D.R, Joachimiak, A, Satchell, K.J.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-02-23 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Albumin-Based Transport of Nonsteroidal Anti-Inflammatory Drugs in Mammalian Blood Plasma.
J.Med.Chem., 2020
|
|
1ED9
 
 | | STRUCTURE OF E. COLI ALKALINE PHOSPHATASE WITHOUT THE INORGANIC PHOSPHATE AT 1.75A RESOLUTION | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Stec, B, Holtz, K.M, Kantrowitz, E.R. | | Deposit date: | 2000-01-27 | | Release date: | 2000-09-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A revised mechanism for the alkaline phosphatase reaction involving three metal ions.
J.Mol.Biol., 299, 2000
|
|
1EO3
 
 | | INHIBITION OF ECORV ENDONUCLEASE BY DEOXYRIBO-3'-S-PHOSPHOROTHIOLATES: A HIGH RESOLUTION X-RAY CRYSTALLOGRAPHIC STUDY | | Descriptor: | ACETIC ACID, DNA (5'-D(*CP*AP*AP*GP*AP*(TSP)P*AP*TP*CP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Horton, N.C, Connolly, B.A, Perona, J.J. | | Deposit date: | 2000-03-21 | | Release date: | 2000-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of EcoRV Endonuclease by Deoxyribo-3'-S-phosphorothiolates: A High-Resolution X-ray Crystallographic Study
J.Am.Chem.Soc., 122, 2000
|
|
5A7P
 
 | | Crystal structure of human JMJD2A in complex with compound 36 | | Descriptor: | 1,2-ETHANEDIOL, 2-[5-[(5-methyl-1,2-oxazol-3-yl)carbonylamino]-2-oxidanyl-phenyl]pyridine-4-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Nowak, R, Velupillai, S, Krojer, T, Gileadi, C, Johansson, C, Korczynska, M, Le, D.D, Younger, N, Gregori-Puigjane, E, Tumber, A, Iwasa, E, Pollock, S.B, Ortiz Torres, I, Kopec, J, Tallant, C, Froese, S, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Shoichet, B.K, Fujimori, D.G, Oppermann, U. | | Deposit date: | 2015-07-09 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Docking and Linking of Fragments to Discover Jumonji Histone Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
