3LMT
 
 | | Crystal structure of DTD from Plasmodium falciparum | | Descriptor: | D-tyrosyl-tRNA(Tyr) deacylase, IODIDE ION | | Authors: | Manickam, Y, Bhatt, T.K, Khan, S, Sharma, A. | | Deposit date: | 2010-02-01 | | Release date: | 2010-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of D-tyrosyl-tRNATyr deacylase using home-source Cu Kalpha and moderate-quality iodide-SAD data: structural polymorphism and HEPES-bound enzyme states
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4WCH
 
 | | Structure of Isolated D Chain of Gigant Hemoglobin from Glossoscolex paulistus | | Descriptor: | Isolated Chain D of Gigant Hemoglobin from Glossoscolex Paulistus, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bachega, J.F.R, Maluf, F.V, Pereira, H.M, Brandao-Neto, J, Tabak, M, Garratt, R.C, Horjales, E. | | Deposit date: | 2014-09-04 | | Release date: | 2015-06-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of the giant haemoglobin from Glossoscolex paulistus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4G2I
 
 | | Structural basis for the accommodation of bis- and tris-aromatic derivatives in Vitamin D Nuclear Receptor | | Descriptor: | (3E,5E)-6-(3-{2-[3,4-bis(hydroxymethyl)phenyl]ethyl}phenyl)-1,1,1-trifluoro-2-(trifluoromethyl)octa-3,5-dien-2-ol, Vitamin D3 receptor | | Authors: | Ciesielski, F, Sato, Y, Moras, D, Rochel, N. | | Deposit date: | 2012-07-12 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the accommodation of bis- and tris-aromatic derivatives in vitamin d nuclear receptor.
J.Med.Chem., 55, 2012
|
|
4W87
 
 | | Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from metagenomic library, in complex with a xyloglucan oligosaccharide | | Descriptor: | MAGNESIUM ION, Xyloglucan-specific endo-beta-1,4-glucanase, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
3V0H
 
 | | Crystal structure of Ciona intestinalis voltage sensor-containing phosphatase (Ci-VSP), residues 241-576(C363S), complexed with D-MYO-inositol-1,4,5-triphosphate | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Voltage-sensor containing phosphatase | | Authors: | Liu, L, Kohout, S.C, Xu, Q, Muller, S, Kimberlin, C, Isacoff, E.Y, Minor, D.L. | | Deposit date: | 2011-12-08 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A glutamate switch controls voltage-sensitive phosphatase function.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4W85
 
 | | Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from ruminal metagenomic library, in complex with glucose | | Descriptor: | MAGNESIUM ION, Xyloglucan-specific endo-beta-1,4-glucanase, beta-D-glucopyranose | | Authors: | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
7SNL
 
 | | Disulfide stabilized HIV-1 CA hexamer in complex with capsid inhibitor N-(1-(3-(4-chloro-1-methyl-3-(methylsulfonamido)-1H-indazol-7-yl)-4-oxo-3,4-dihydropyrido[2,3-d]pyrimidin-2-yl)-2-(3,5-difluorophenyl)ethyl)-2-(3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl)acetamide | | Descriptor: | CHLORIDE ION, Capsid protein p24, IODIDE ION, ... | | Authors: | Bester, S.M, Kvaratskhelia, M. | | Deposit date: | 2021-10-28 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Disulfide stabilized HIV-1 CA hexamer in complex with capsid inhibitor N-(1-(3-(4-chloro-1-methyl-3-(methylsulfonamido)-1H-indazol-7-yl)-4-oxo-3,4-dihydropyrido[2,3-d]pyrimidin-2-yl)-2-(3,5-difluorophenyl)ethyl)-2-(3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl)acetamide
To Be Published
|
|
4UOI
 
 | | Unexpected structure for the N-terminal domain of Hepatitis C virus envelope glycoprotein E1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GENOME POLYPROTEIN | | Authors: | El Omari, K, Iourin, O, Kadlec, J, Harlos, K, Grimes, J.M, Stuart, D.I. | | Deposit date: | 2014-06-04 | | Release date: | 2014-08-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Unexpected Structure for the N-Terminal Domain of Hepatitis C Virus Envelope Glycoprotein E1
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5XHW
 
 | | Crystal structure of HddC from Yersinia pseudotuberculosis | | Descriptor: | Putative 6-deoxy-D-mannoheptose pathway protein, SULFATE ION | | Authors: | Park, J, Kim, H, Kim, S, Shin, D.H. | | Deposit date: | 2017-04-24 | | Release date: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of d-glycero-alpha-d-manno-heptose-1-phosphate guanylyltransferase from Yersinia pseudotuberculosis.
Biochim. Biophys. Acta, 1866, 2018
|
|
6N2H
 
 | | Structure of D-ornithine/D-lysine decarboxylase from Salmonella typhimurium | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, D-ornithine/D-lysine decarboxylase, DIMETHYL SULFOXIDE | | Authors: | Phillips, R.S, Hoover, T.R. | | Deposit date: | 2018-11-13 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal Structure of d-Ornithine/d-Lysine Decarboxylase, a Stereoinverting Decarboxylase: Implications for Substrate Specificity and Stereospecificity of Fold III Decarboxylases.
Biochemistry, 58, 2019
|
|
6XXG
 
 | | Structure of truncated 1-deoxy-D-xylulose 5-phosphate synthase (DXS) from Deinococcus radiodurans | | Descriptor: | 1-deoxy-D-xylulose-5-phosphate synthase,1-deoxy-D-xylulose-5-phosphate synthase, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Gierse, R.M, Reddem, E, Grooves, M.R. | | Deposit date: | 2020-01-27 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Identification of a 1-deoxy-D-xylulose-5-phosphate synthase (DXS) mutant with improved crystallographic properties.
Biochem.Biophys.Res.Commun., 539, 2021
|
|
8IDN
 
 | | Cryo-EM structure of RBD/E77-Fab complex | | Descriptor: | E77 Fab heavy chain, E77 Fab light chain, Spike protein S1, ... | | Authors: | Lu, D.F, Zhang, Z.C. | | Deposit date: | 2023-02-13 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | The structure of the RBD-E77 Fab complex reveals neutralization and immune escape of SARS-CoV-2.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6N1E
 
 | | Crystal structure of X. citri phosphoglucomutase in complex with 1-methyl-glucose 6-phosphate | | Descriptor: | 1-deoxy-7-O-phosphono-alpha-D-gluco-hept-2-ulopyranose, MAGNESIUM ION, Phosphomannomutase/phosphoglucomutase | | Authors: | Beamer, L.J, Stiers, K.M. | | Deposit date: | 2018-11-08 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis, Derivatization, and Structural Analysis of Phosphorylated Mono-, Di-, and Trifluorinated d-Gluco-heptuloses by Glucokinase: Tunable Phosphoglucomutase Inhibition.
Acs Omega, 4, 2019
|
|
5XNI
 
 | |
6WYZ
 
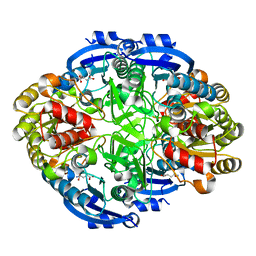 | | Crystal structure of Pseudomonas 7A Glutaminase-Asparaginase (mutant K173M) in complex with D-Glu at pH 5.5 | | Descriptor: | D-GLUTAMIC ACID, Glutaminase-asparaginase | | Authors: | Strzelczyk, P, Zhang, D, Wlodawer, A, Lubkowski, J. | | Deposit date: | 2020-05-13 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Generalized enzymatic mechanism of catalysis by tetrameric L-asparaginases from mesophilic bacteria.
Sci Rep, 10, 2020
|
|
6X0S
 
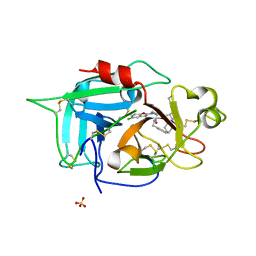 | |
6X0T
 
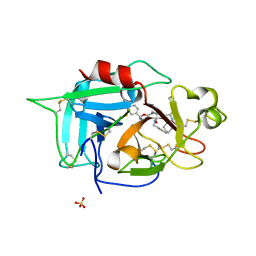 | | Structure of human plasma factor XIIa in complex with (2S)-1-(N,3-dicyclohexyl-D-alanyl)-4-[(4R,5S)-4-methyl-5-phenyl-4,5-dihydro-1,3-oxazol-2-yl]-N-[(thiophen-2-yl)methyl]piperazine-2-carboxamide (compound 8h) | | Descriptor: | (2S)-1-(N,3-dicyclohexyl-D-alanyl)-4-[(4R,5S)-4-methyl-5-phenyl-4,5-dihydro-1,3-oxazol-2-yl]-N-[(thiophen-2-yl)methyl]piperazine-2-carboxamide, Coagulation factor XII, SULFATE ION | | Authors: | Orth, P. | | Deposit date: | 2020-05-17 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.388 Å) | | Cite: | Structure of human plasma factor XIIa in complex with (2S)-1-(N,3-dicyclohexyl-D-alanyl)-4-[(4R,5S)-4-methyl-5-phenyl-4,5-dihydro-1,3-oxazol-2-yl]-N-[(thiophen-2-yl)methyl]piperazine-2-carboxamide (compound 8h)
To Be Published
|
|
8BD0
 
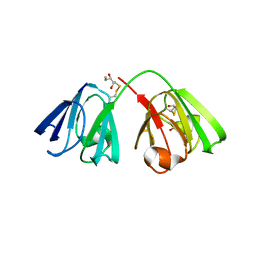 | |
8BPI
 
 | |
6ZN1
 
 | | Trehalose transferase bound to alpha-D-glucopyranosyl-beta-galactopyranose from Thermoproteus uzoniensis | | Descriptor: | DI(HYDROXYETHYL)ETHER, THIOCYANATE ION, Trehalose phosphorylase/synthase, ... | | Authors: | Bento, I, Mestrom, L, Marsden, S.R, van der Eijk, H, Laustsen, J.U, Jeffries, C.M, Svergun, D.I, Hagedoorn, P.-H, Hanefeld, U. | | Deposit date: | 2020-07-06 | | Release date: | 2021-01-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Anomeric Selectivity of Trehalose Transferase with Rare l-Sugars.
ACS Catal, 10, 2020
|
|
8RZK
 
 | | The Michaelis complex of ZgGH129 D486N from Zobellia galactanivorans with neo-b/k-oligo-carrageenan tetrasaccharide (beta-kappa neo-oligo-carrageenan DP4). | | Descriptor: | 1,2-ETHANEDIOL, 3,6-anhydro-alpha-D-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose, CHLORIDE ION, ... | | Authors: | Roret, T, Czjzek, M, Ficko-Blean, E. | | Deposit date: | 2024-02-12 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Constrained Catalytic Itinerary of a Retaining 3,6-Anhydro-D-Galactosidase, a Key Enzyme in Red Algal Cell Wall Degradation.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8BZJ
 
 | | Human MST3 (STK24) kinase in complex with inhibitor MRLW5 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-6-[2-chloranyl-4-(6-methylpyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Balourdas, D.I, Rak, M, Tesch, R, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-12-14 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Development of Selective Pyrido[2,3- d ]pyrimidin-7(8 H )-one-Based Mammalian STE20-Like (MST3/4) Kinase Inhibitors.
J.Med.Chem., 67, 2024
|
|
8HPC
 
 | |
7A9G
 
 | | Truncated 1-deoxy-D-xylulose 5-phosphate synthase (DXS) from Mycobacterium tuberculosis with intermediate 2-acetyl-thiamine diphosphate | | Descriptor: | 1-deoxy-D-xylulose-5-phosphate synthase,1-deoxy-D-xylulose-5-phosphate synthase, 2-ACETYL-THIAMINE DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gierse, R.M, Reddem, E, Grooves, M.R. | | Deposit date: | 2020-09-02 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | First crystal structures of 1-deoxy-D-xylulose 5-phosphate synthase (DXPS) from Mycobacterium tuberculosis indicate a distinct mechanism of intermediate stabilization.
Sci Rep, 12, 2022
|
|
7A9H
 
 | | Truncated 1-deoxy-D-xylulose 5-phosphate synthase (DXS) from Mycobacterium tuberculosis | | Descriptor: | 1-deoxy-D-xylulose-5-phosphate synthase,1-deoxy-D-xylulose-5-phosphate synthase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Gierse, R.M, Reddem, E, Grooves, M.R. | | Deposit date: | 2020-09-02 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | First crystal structures of 1-deoxy-D-xylulose 5-phosphate synthase (DXPS) from Mycobacterium tuberculosis indicate a distinct mechanism of intermediate stabilization.
Sci Rep, 12, 2022
|
|
