3OC8
 
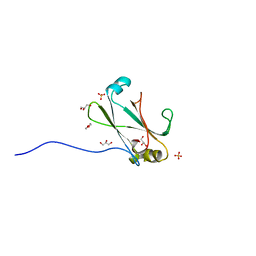 | | Crystal Structure of the C-terminal Domain of the Vibrio cholerae soluble colonization factor TcpF | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Craig, L, Kolappan, S, Yuen, A.S.W. | | Deposit date: | 2010-08-09 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Vibrio cholerae Colonization Factor TcpF and Identification of a Functional Immunogenic Site.
J.Mol.Biol., 409, 2011
|
|
3OCC
 
 | | Crystal structure of PNP with DADMEimmH from Yersinia pseudotuberculosis | | Descriptor: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, Purine nucleoside phosphorylase deoD-type | | Authors: | Kim, J, Ramagopal, U.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-08-09 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | crystal structure of PNP with DADMEimmH from Yersinia pseudotuberculosis
To be Published
|
|
1UEQ
 
 | | Solution Structure of The First PDZ domain of Human Atrophin-1 Interacting Protein 1 (KIAA0705 protein) | | Descriptor: | MEMBRANE ASSOCIATED GUANYLATE KINASE INVERTED-2 (MAGI-2) | | Authors: | Zhao, C, Kigawa, T, Tochio, N, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-20 | | Release date: | 2003-11-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of The First PDZ domain of Human Atrophin-1 Interacting Protein 1 (KIAA0705 protein)
To be Published
|
|
2D7C
 
 | | Crystal structure of human Rab11 in complex with FIP3 Rab-binding domain | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shiba, T, Koga, H, Shin, H.W, Kawasaki, M, Kato, R, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2005-11-16 | | Release date: | 2006-09-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for Rab11-dependent membrane recruitment of a family of Rab11-interacting protein 3 (FIP3)/Arfophilin-1.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1BC2
 
 | | ZN-DEPENDENT METALLO-BETA-LACTAMASE FROM BACILLUS CEREUS | | Descriptor: | METALLO-BETA-LACTAMASE II, SULFATE ION, ZINC ION | | Authors: | Fabiane, S.M, Sutton, B.J. | | Deposit date: | 1997-04-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the zinc-dependent beta-lactamase from Bacillus cereus at 1.9 A resolution: binuclear active site with features of a mononuclear enzyme.
Biochemistry, 37, 1998
|
|
3OF8
 
 | |
3CHV
 
 | |
4I0D
 
 | |
1AVS
 
 | |
4HZC
 
 | |
1UAI
 
 | |
6M18
 
 | | ACE2-B0AT1 complex | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Xia, L, Zhou, Q. | | Deposit date: | 2020-02-25 | | Release date: | 2020-03-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2.
Science, 367, 2020
|
|
1P0Q
 
 | | Crystal structure of soman-aged human butyryl cholinesterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Nicolet, Y, Lockridge, O, Masson, P, Fontecilla-Camps, J.C, Nachon, F. | | Deposit date: | 2003-04-10 | | Release date: | 2003-08-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal structure of human butyrylcholinesterase and of its complexes with substrate and products.
J.Biol.Chem., 278, 2003
|
|
3OKA
 
 | | Crystal structure of Corynebacterium glutamicum PimB' in complex with GDP-Man (triclinic crystal form) | | Descriptor: | GDP-mannose-dependent alpha-(1-6)-phosphatidylinositol monomannoside mannosyltransferase, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, ... | | Authors: | Batt, S.M, Jabeen, T, Besra, G.S, Futterer, K. | | Deposit date: | 2010-08-24 | | Release date: | 2010-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Acceptor substrate discrimination in phosphatidyl-myo-inositol mannoside synthesis: structural and mutational analysis of mannosyltransferase Corynebacterium glutamicum PimB'.
J.Biol.Chem., 285, 2010
|
|
1IEM
 
 | | Crystal Structure of AmpC beta-lactamase from E. coli in Complex with a Boronic Acid Inhibitor (1, CefB4) | | Descriptor: | PHOSPHATE ION, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE, beta-lactamase | | Authors: | Powers, R.A, Caselli, E, Focia, P.J, Prati, F, Shoichet, B.K. | | Deposit date: | 2001-04-10 | | Release date: | 2001-08-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of ceftazidime and its transition-state analogue in complex with AmpC beta-lactamase: implications for resistance mutations and inhibitor design.
Biochemistry, 40, 2001
|
|
1DYE
 
 | |
2M2Q
 
 | | Solution structure of MCh-1: A novel inhibitor cystine knot peptide from Momordica charantia | | Descriptor: | Inhibitor cystine knot peptide MCh-1 | | Authors: | He, W, Chan, L, Clark, R.J, Tang, J, Zeng, G, Franco, O.L, Cantacessi, C, Craik, D.J, Daly, N.L, Tan, N. | | Deposit date: | 2013-01-01 | | Release date: | 2013-11-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Novel Inhibitor Cystine Knot Peptides from Momordica charantia.
Plos One, 8, 2013
|
|
4KS6
 
 | | Crystal structure of the catalytic domain of botulinum neurotoxin BoNT/A C134S mutant with covalent inhibitor that modifies Cys-165 causing disorder in 166-174 stretch | | Descriptor: | 1,2-ETHANEDIOL, Botulinum neurotoxin A light chain, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stura, E.A, Vera, L, Guitot, K, Dive, V. | | Deposit date: | 2013-05-17 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Covalent modification of the active site cysteine stresses Clostridium botulinum neurotoxin A
To be Published
|
|
2GN0
 
 | |
1DYA
 
 | |
1DYG
 
 | |
2Z4T
 
 | | Crystal Structure of Vibrionaceae Photobacterium sp. JT-ISH-224 2,6-sialyltransferase in a Ternary Complex with Donor Product CMP and Accepter Substrate Lactose | | Descriptor: | Beta-galactoside alpha-2,6-sialyltransferase, CYTIDINE-5'-MONOPHOSPHATE, GLYCEROL, ... | | Authors: | Kakuta, Y, Okino, N, Kajiwara, H, Ichikawa, M, Takakura, Y, Ito, M, Yamamoto, T. | | Deposit date: | 2007-06-25 | | Release date: | 2008-04-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Vibrionaceae Photobacterium sp. JT-ISH-224 alpha2,6-sialyltransferase in a ternary complex with donor product CMP and acceptor substrate lactose: catalytic mechanism and substrate recognition
Glycobiology, 18, 2008
|
|
2CES
 
 | | Beta-glucosidase from Thermotoga maritima in complex with glucoimidazole | | Descriptor: | ACETATE ION, BETA-GLUCOSIDASE A, CALCIUM ION, ... | | Authors: | Gloster, T.M, Roberts, S, Vasella, A, Davies, G.J. | | Deposit date: | 2006-02-10 | | Release date: | 2006-09-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural, Kinetic, and Thermodynamic Analysis of Glucoimidazole-Derived Glycosidase Inhibitors.
Biochemistry, 45, 2006
|
|
3SFC
 
 | | Structure-Based Optimization of Potent 4- and 6-Azaindole-3-Carboxamides as Renin Inhibitors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Renin, ... | | Authors: | Scheiper, B, Matter, H, Steinhagen, H, Bocskei, Z, Fleury, V, McCort, G. | | Deposit date: | 2011-06-13 | | Release date: | 2011-08-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based optimization of potent 4- and 6-azaindole-3-carboxamides as renin inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1XXO
 
 | | X-ray crystal structure of mycobacterium tuberculosis pyridoxine 5'-phosphate oxidase at 1.8 a resolution | | Descriptor: | hypothetical protein Rv1155 | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Garen, C, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-11-07 | | Release date: | 2004-11-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of mycobacterium tuberculosis pyridoxine 5'-phosphate oxidase at 1.8 a resolution
To be Published
|
|
