8CRE
 
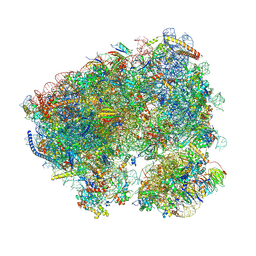 | | Crystal structure of the Candida albicans 80S ribosome in complex with geneticin G418 | | Descriptor: | 18S, 25S, 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Kolosova, O, Zgadzay, Y, Yusupov, M. | | Deposit date: | 2023-03-08 | | Release date: | 2024-09-18 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanism of read-through enhancement by aminoglycosides and mefloquine
Proc.Natl.Acad.Sci.USA, 2025
|
|
1RHY
 
 | | Crystal structure of Imidazole Glycerol Phosphate Dehydratase | | Descriptor: | ACETIC ACID, ETHYL MERCURY ION, GLYCEROL, ... | | Authors: | Sinha, S.C, Chaudhuri, B.N, Burgner, J.W, Yakovleva, G, Davisson, V.J, Smith, J.L. | | Deposit date: | 2003-11-14 | | Release date: | 2004-05-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of imidazole glycerol-phosphate dehydratase: duplication of an unusual fold
J.Biol.Chem., 279, 2004
|
|
2I9X
 
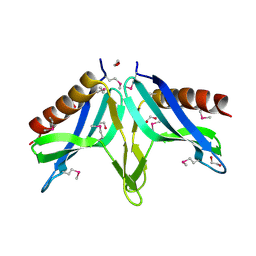 | | Structural Genomics, the crystal structure of SpoVG conserved domain from Staphylococcus epidermidis ATCC 12228 | | Descriptor: | 1,2-ETHANEDIOL, Putative septation protein spoVG | | Authors: | Tan, K, Maltseva, N, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-06 | | Release date: | 2006-10-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of SpoVG from Staphylococcus epidermidis ATCC 12228
To be Published
|
|
2C5D
 
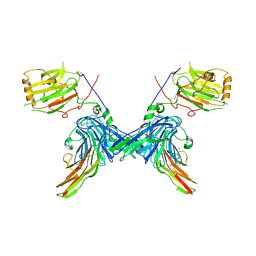 | | Structure of a minimal Gas6-Axl complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GROWTH-ARREST-SPECIFIC PROTEIN 6 PRECURSOR, ... | | Authors: | Sasaki, T, Knyazev, P.G, Clout, N.J, Cheburkin, Y, Goehring, W, Ullrich, A, Timpl, R, Hohenester, E. | | Deposit date: | 2005-10-26 | | Release date: | 2005-12-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Basis for Gas6-Axl Signalling.
Embo J., 25, 2006
|
|
6EUF
 
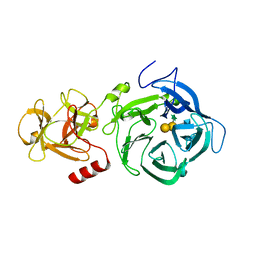 | | The GH43, Beta 1,3 Galactosidase, BT0265 | | Descriptor: | Beta-glucanase, alpha-L-arabinofuranose-(1-3)-[alpha-L-arabinofuranose-(1-4)][beta-D-glucopyranuronic acid-(1-6)]beta-D-galactopyranose-(1-6)-beta-D-galactopyranose, alpha-L-rhamnopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-6)-[alpha-L-arabinofuranose-(1-3)][alpha-L-arabinofuranose-(1-4)]beta-D-galactopyranose-(1-6)-beta-D-galactopyranose | | Authors: | Cartmell, A, Gilbert, H.J. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation.
Nat Microbiol, 3, 2018
|
|
1VMJ
 
 | |
1YYL
 
 | | crystal structure of CD4M33, a scorpion-toxin mimic of CD4, in complex with HIV-1 YU2 gp120 envelope glycoprotein and anti-HIV-1 antibody 17b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD4M33, scorpion-toxin mimic of CD4, ... | | Authors: | Huang, C.C, Stricher, F, Martin, L, Decker, J.M, Majeed, S, Barthe, P, Hendrickson, W.A, Robinson, J, Roumestand, C, Sodroski, J, Wyatt, R, Shaw, G.M, Vita, C, Kwong, P.D. | | Deposit date: | 2005-02-25 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Scorpion-toxin mimics of CD4 in complex with human immunodeficiency virus gp120 crystal structures, molecular mimicry, and neutralization breadth.
Structure, 13, 2005
|
|
4LJZ
 
 | | Crystal Structure Analysis of the E.coli holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Bae, B, Darst, S.A. | | Deposit date: | 2013-07-05 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5871 Å) | | Cite: | Phage T7 Gp2 inhibition of Escherichia coli RNA polymerase involves misappropriation of sigma 70 domain 1.1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4FIX
 
 | | Crystal Structure of GlfT2 | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Wheatley, R.W, Zheng, R.B, Lowary, T.L, Ng, K.K.S. | | Deposit date: | 2012-06-11 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Tetrameric Structure of the GlfT2 Galactofuranosyltransferase Reveals a Scaffold for the Assembly of Mycobacterial Arabinogalactan.
J.Biol.Chem., 287, 2012
|
|
3GXP
 
 | | Crystal structure of acid-alpha-galactosidase A complexed with galactose at pH 4.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-galactosidase A, ... | | Authors: | Lieberman, R.L. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Effects of pH and iminosugar pharmacological chaperones on lysosomal glycosidase structure and stability.
Biochemistry, 48, 2009
|
|
4FRS
 
 | |
6EZ9
 
 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) - the E424M inactive mutant, in complex with a inhibitor JHU3372 | | Descriptor: | (2~{S})-2-[[(2~{S})-4-methyl-1-oxidanyl-1-oxidanylidene-pentan-2-yl]oxycarbonylamino]pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Barinka, C, Novakova, Z, Motlova, L. | | Deposit date: | 2017-11-14 | | Release date: | 2018-12-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural and computational basis for potent inhibition of glutamate carboxypeptidase II by carbamate-based inhibitors.
Bioorg.Med.Chem., 27, 2019
|
|
1RF2
 
 | | Cholera Toxin B-Pentamer Complexed With Bivalent Nitrophenol-Galactoside Ligand BV4 | | Descriptor: | 1,3-BIS-([3-[3-[3-(4-{3-[3-NITRO-5-(GALACTOPYRANOSYLOXY)-BENZOYLAMINO]-PROPYL}-PIPERAZIN-1-YL)-PROPYLAMINO-3,4-DIOXO-CYCLOBUTENYL]-AMINO-PROPOXY-ETHOXY-ETHOXY]-PROPYL-]AMINO-CARBONYLOXY)-2-AMINO-PROPANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, TRIETHYLENE GLYCOL, ... | | Authors: | Pickens, J.C, Mitchell, D.D, Liu, J, Tan, X, Zhang, Z, Verlinde, C.L, Hol, W.G, Fan, E. | | Deposit date: | 2003-11-07 | | Release date: | 2004-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Nonspanning bivalent ligands as improved surface receptor binding inhibitors of the cholera toxin B pentamer.
Chem.Biol., 11, 2004
|
|
2HX0
 
 | | Three-dimensional structure of the hypothetical protein from Salmonella cholerae-suis (aka Salmonella enterica) at the resolution 1.55 A. Northeast Structural Genomics target ScR59. | | Descriptor: | MAGNESIUM ION, Putative DNA-binding protein | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Shastry, R, Conover, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-08-02 | | Release date: | 2006-09-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Three-dimensional structure of the hypothetical protein from Salmonella cholerae-suis (aka Salmonella enterica) at the resolution 1.55 A. Northeast Structural Genomics target ScR59.
To be Published
|
|
2HY3
 
 | | Crystal structure of the human tyrosine receptor phosphate gamma in complex with vanadate | | Descriptor: | Receptor-type tyrosine-protein phosphatase gamma, VANADATE ION | | Authors: | Jin, X, Min, T, Bera, A, Mu, H, Sauder, J.M, Freeman, J.C, Reyes, C, Smith, D, Wasserman, S.R, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-04 | | Release date: | 2006-09-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2FYF
 
 | | Structure of a putative phosphoserine aminotransferase from Mycobacterium Tuberculosis | | Descriptor: | GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION, ... | | Authors: | Coulibaly, F, Lassalle, E, Baker, E.N, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2006-02-07 | | Release date: | 2007-01-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of phosphoserine aminotransferase from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5POP
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with N10987a | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(pyridin-4-yl)piperazin-1-yl]ethan-1-one, Bromodomain-containing protein 1, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.579 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
1RJN
 
 | | The Crystal Structure of MenB (Rv0548c) from Mycobacterium tuberculosis in Complex with the CoA Portion of Naphthoyl CoA | | Descriptor: | 3-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]PROPANE-1-SULFONIC ACID, COENZYME A, menB | | Authors: | Johnston, J.M, Arcus, V.L, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-11-19 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of naphthoate synthase (MenB) from Mycobacterium tuberculosis in both native and product-bound forms.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2FZ5
 
 | | Solution structure of two-electron reduced Megasphaera elsdenii flavodoxin | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, Flavodoxin | | Authors: | van Mierlo, C.P.M, Lijnzaad, P, Vervoort, J, Mueller, F, Berendsen, H.J, de Vlieg, J. | | Deposit date: | 2006-02-09 | | Release date: | 2006-03-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Tertiary structure of two-electron reduced Megasphaera elsdenii flavodoxin and some implications, as determined by two-dimensional 1H NMR and restrained molecular dynamics
Eur.J.Biochem., 194, 1990
|
|
2FYS
 
 | | Crystal structure of Erk2 complex with KIM peptide derived from MKP3 | | Descriptor: | Dual specificity protein phosphatase 6, Mitogen-activated protein kinase 1 | | Authors: | Liu, S, Sun, J.P, Zhou, B, Zhang, Z.Y. | | Deposit date: | 2006-02-08 | | Release date: | 2006-04-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of docking interactions between ERK2 and MAP kinase phosphatase 3
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2ZJH
 
 | |
3OFU
 
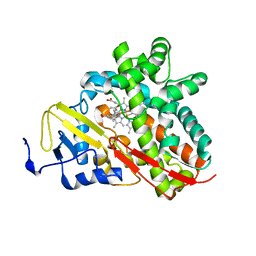 | | Crystal Structure of Cytochrome P450 CYP101C1 | | Descriptor: | (3E)-4-(2,6,6-trimethylcyclohex-1-en-1-yl)but-3-en-2-one, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zhou, W, Ma, M, Bell, S.G, Yang, W, Hao, Y, Rees, N.H, Bartlam, M, Wong, L.-L, Rao, Z. | | Deposit date: | 2010-08-16 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Analysis of CYP101C1 from Novosphingobium aromaticivorans DSM12444.
Chembiochem, 12, 2011
|
|
2ZJN
 
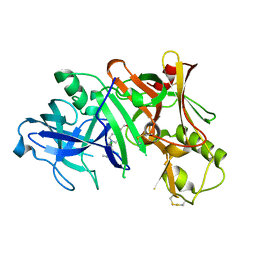 | |
5EAH
 
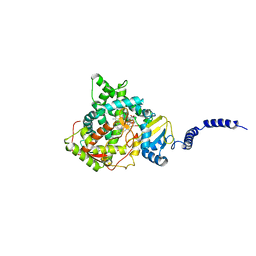 | | Saccharomyces cerevisiae CYP51 complexed with the plant pathogen inhibitor Difenoconazole | | Descriptor: | 1-[[(2~{R},4~{R})-2-[2-chloranyl-4-(4-chloranylphenoxy)phenyl]-4-methyl-1,3-dioxolan-2-yl]methyl]-1,2,4-triazole, 1-[[(2~{R},4~{S})-2-[2-chloranyl-4-(4-chloranylphenoxy)phenyl]-4-methyl-1,3-dioxolan-2-yl]methyl]-1,2,4-triazole, 1-[[(2~{S},4~{R})-2-[2-chloranyl-4-(4-chloranylphenoxy)phenyl]-4-methyl-1,3-dioxolan-2-yl]methyl]-1,2,4-triazole, ... | | Authors: | Tyndall, J.D.A, Sabherwal, M, Keniya, M.V, Wilson, R.K, Woods, M.V, Monk, B.C. | | Deposit date: | 2015-10-16 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.541 Å) | | Cite: | Structural and Functional Elucidation of Yeast Lanosterol 14 alpha-Demethylase in Complex with Agrochemical Antifungals.
PLoS ONE, 11, 2016
|
|
1AAY
 
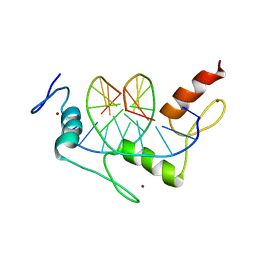 | | ZIF268 ZINC FINGER-DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*CP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*GP*CP*CP*CP*AP*CP*GP*C)-3'), PROTEIN (ZIF268 ZINC FINGER PEPTIDE), ... | | Authors: | Elrod-Erickson, M, Rould, M.A, Pabo, C.O. | | Deposit date: | 1997-01-18 | | Release date: | 1997-04-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Zif268 protein-DNA complex refined at 1.6 A: a model system for understanding zinc finger-DNA interactions.
Structure, 4, 1996
|
|
